Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
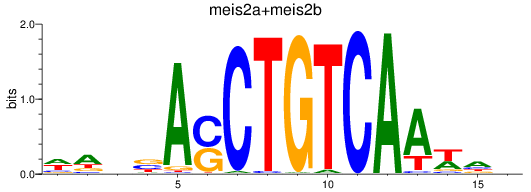
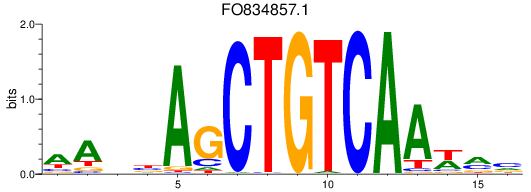
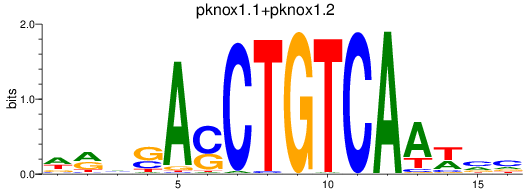
Results for meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Z-value: 0.69
Transcription factors associated with meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis2b
|
ENSDARG00000077840 | Meis homeobox 2a |
|
meis2a
|
ENSDARG00000098240 | Meis homeobox 2a |
|
FO834857.1
|
ENSDARG00000112895 | homeobox protein TGIF2-like |
|
pknox1.1
|
ENSDARG00000018765 | pbx/knotted 1 homeobox 1.1 |
|
pknox1.2
|
ENSDARG00000036542 | pbx/knotted 1 homeobox 1.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FO834857.1 | dr11_v1_chr6_-_1820606_1820606 | 0.83 | 1.9e-05 | Click! |
| meis2b | dr11_v1_chr20_-_10118818_10118818 | -0.67 | 2.3e-03 | Click! |
| meis2a | dr11_v1_chr17_+_52822831_52822831 | -0.66 | 3.0e-03 | Click! |
| pknox1.2 | dr11_v1_chr1_-_46981134_46981134 | -0.65 | 3.4e-03 | Click! |
| pknox1.1 | dr11_v1_chr9_+_19529951_19529951 | 0.31 | 2.1e-01 | Click! |
Activity profile of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
Sorted Z-values of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_32385989 | 2.65 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr2_-_11027258 | 2.10 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr12_-_48168135 | 1.81 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr4_-_20081621 | 1.80 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr14_-_41468892 | 1.74 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr3_+_7771420 | 1.48 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr5_-_12587053 | 1.48 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr20_+_14968031 | 1.45 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr22_-_37834312 | 1.38 |
ENSDART00000076128
|
ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr25_+_22319940 | 1.38 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr5_-_32489796 | 1.35 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr1_+_2301961 | 1.34 |
ENSDART00000108919
ENSDART00000143361 ENSDART00000142944 |
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr17_-_7792376 | 1.33 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr8_+_25959940 | 1.32 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr23_+_13528053 | 1.28 |
ENSDART00000162217
|
uckl1b
|
uridine-cytidine kinase 1-like 1b |
| chr15_-_28587490 | 1.27 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr5_-_69482891 | 1.25 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr15_-_28587147 | 1.23 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr7_-_26262978 | 1.20 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr7_-_26263183 | 1.19 |
ENSDART00000079357
ENSDART00000190369 ENSDART00000193154 ENSDART00000101109 |
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr4_+_27898833 | 1.12 |
ENSDART00000146099
|
cerk
|
ceramide kinase |
| chr11_-_10456387 | 1.10 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr12_+_33320884 | 1.10 |
ENSDART00000188988
|
csnk1db
|
casein kinase 1, delta b |
| chr25_-_2723682 | 1.06 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr18_+_39487486 | 1.06 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr9_+_27876146 | 1.02 |
ENSDART00000133997
|
armc8
|
armadillo repeat containing 8 |
| chr14_-_34605607 | 1.00 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr6_-_41135215 | 1.00 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr23_+_21492151 | 1.00 |
ENSDART00000025487
|
icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr24_-_36095526 | 1.00 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr2_-_27651674 | 0.99 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr19_-_30800004 | 0.99 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr23_+_39963599 | 0.97 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr10_+_28160265 | 0.97 |
ENSDART00000022484
|
rnft1
|
ring finger protein, transmembrane 1 |
| chr5_-_13766651 | 0.96 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr14_+_30340251 | 0.94 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr5_-_23675222 | 0.94 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr21_-_14832369 | 0.91 |
ENSDART00000144859
|
pus1
|
pseudouridylate synthase 1 |
| chr21_-_41065369 | 0.89 |
ENSDART00000143749
|
larsb
|
leucyl-tRNA synthetase b |
| chr18_+_35128685 | 0.89 |
ENSDART00000151579
|
si:ch211-195m9.3
|
si:ch211-195m9.3 |
| chr14_-_6402769 | 0.89 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr5_-_39805620 | 0.89 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr5_+_27137473 | 0.87 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr15_+_41815703 | 0.86 |
ENSDART00000059508
|
pxylp1
|
2-phosphoxylose phosphatase 1 |
| chr17_+_19630068 | 0.85 |
ENSDART00000182619
|
rgs7a
|
regulator of G protein signaling 7a |
| chr23_+_33963619 | 0.85 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr13_+_46803979 | 0.84 |
ENSDART00000159260
|
CU695232.1
|
|
| chr5_-_41124241 | 0.82 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr14_-_33481428 | 0.80 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr14_-_34605804 | 0.79 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr14_-_38889840 | 0.79 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr19_-_46032556 | 0.77 |
ENSDART00000167185
|
nup153
|
nucleoporin 153 |
| chr17_-_23673864 | 0.77 |
ENSDART00000104738
ENSDART00000128958 |
ptena
|
phosphatase and tensin homolog A |
| chr5_-_39805874 | 0.76 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr25_-_36261836 | 0.76 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr7_-_56766100 | 0.75 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr8_-_410728 | 0.74 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr11_-_34478225 | 0.72 |
ENSDART00000189604
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr16_+_33144306 | 0.71 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_-_33945692 | 0.70 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr16_+_50434668 | 0.68 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr12_+_28888975 | 0.68 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr10_+_36695597 | 0.67 |
ENSDART00000169015
ENSDART00000171392 |
rab6a
|
RAB6A, member RAS oncogene family |
| chr23_-_31060350 | 0.66 |
ENSDART00000145598
ENSDART00000191491 |
si:ch211-197l9.5
|
si:ch211-197l9.5 |
| chr10_-_35177257 | 0.66 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr17_+_49095662 | 0.65 |
ENSDART00000179545
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr19_+_42231431 | 0.65 |
ENSDART00000102698
|
jtb
|
jumping translocation breakpoint |
| chr8_-_18899427 | 0.64 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr19_-_12212692 | 0.64 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr17_-_24890843 | 0.64 |
ENSDART00000184984
ENSDART00000135569 ENSDART00000193661 |
gale
|
UDP-galactose-4-epimerase |
| chr18_+_14619544 | 0.64 |
ENSDART00000010821
|
utp4
|
UTP4, small subunit processome component |
| chr18_-_46010 | 0.63 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr16_-_51072406 | 0.63 |
ENSDART00000083777
|
ago3a
|
argonaute RISC catalytic component 3a |
| chr3_+_29082267 | 0.63 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr6_+_33537267 | 0.63 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr15_+_23528010 | 0.63 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr8_-_4760723 | 0.62 |
ENSDART00000064201
|
cdc45
|
CDC45 cell division cycle 45 homolog (S. cerevisiae) |
| chr5_-_20135679 | 0.61 |
ENSDART00000079402
|
usp30
|
ubiquitin specific peptidase 30 |
| chr13_-_24396003 | 0.60 |
ENSDART00000016211
|
tbp
|
TATA box binding protein |
| chr8_-_13678415 | 0.60 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr10_-_25328814 | 0.60 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr10_+_10738880 | 0.60 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr3_-_40836081 | 0.60 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr15_-_20528494 | 0.60 |
ENSDART00000048423
|
timm50
|
translocase of inner mitochondrial membrane 50 homolog (S. cerevisiae) |
| chr6_+_21005725 | 0.58 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr18_-_41160975 | 0.58 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr9_+_34151367 | 0.58 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr20_-_28433990 | 0.58 |
ENSDART00000182824
ENSDART00000193381 |
wdr21
|
WD repeat domain 21 |
| chr9_+_6009077 | 0.56 |
ENSDART00000057484
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr22_-_21897203 | 0.56 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr8_-_41519064 | 0.56 |
ENSDART00000098578
ENSDART00000112214 |
golga1
|
golgin A1 |
| chr22_+_18316144 | 0.56 |
ENSDART00000137985
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr20_+_27712714 | 0.55 |
ENSDART00000008306
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr13_-_24396199 | 0.55 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr23_-_29667544 | 0.54 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr20_+_27713210 | 0.54 |
ENSDART00000132222
|
zbtb1
|
zinc finger and BTB domain containing 1 |
| chr22_-_37796998 | 0.54 |
ENSDART00000124742
ENSDART00000191232 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr8_-_41519234 | 0.53 |
ENSDART00000167283
ENSDART00000180666 |
golga1
|
golgin A1 |
| chr20_+_21391181 | 0.53 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr7_+_41887429 | 0.53 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr22_+_2769236 | 0.53 |
ENSDART00000141836
|
si:dkey-20i20.10
|
si:dkey-20i20.10 |
| chr15_-_506010 | 0.52 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr17_-_14780578 | 0.52 |
ENSDART00000154690
|
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr20_-_165818 | 0.51 |
ENSDART00000123860
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr15_-_20125331 | 0.50 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr13_+_24396666 | 0.50 |
ENSDART00000139197
ENSDART00000101200 |
zgc:153169
|
zgc:153169 |
| chr22_-_17677947 | 0.49 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr16_+_33144112 | 0.49 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr15_+_17345609 | 0.49 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr12_+_18916285 | 0.48 |
ENSDART00000127536
|
cbx7b
|
chromobox homolog 7b |
| chr10_+_20590190 | 0.47 |
ENSDART00000131819
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_+_9612574 | 0.47 |
ENSDART00000150336
ENSDART00000041289 ENSDART00000150828 |
tmem243b
|
transmembrane protein 243, mitochondrial b |
| chr5_+_44346691 | 0.47 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr3_+_28576173 | 0.47 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr14_+_29200772 | 0.47 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr16_-_32672883 | 0.47 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr2_+_2957515 | 0.46 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr22_-_16377666 | 0.45 |
ENSDART00000161878
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr5_-_37103487 | 0.45 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr21_-_25612658 | 0.45 |
ENSDART00000115276
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr16_-_44673851 | 0.45 |
ENSDART00000015139
|
dcaf13
|
ddb1 and cul4 associated factor 13 |
| chr16_+_20167811 | 0.44 |
ENSDART00000004031
|
hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr17_-_50040927 | 0.44 |
ENSDART00000184304
|
FO834825.1
|
|
| chr23_-_4019928 | 0.44 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr15_+_23528310 | 0.44 |
ENSDART00000152523
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr17_+_50074372 | 0.44 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr22_-_16377960 | 0.44 |
ENSDART00000168170
|
ttc39c
|
tetratricopeptide repeat domain 39C |
| chr6_+_38773376 | 0.43 |
ENSDART00000078128
ENSDART00000184053 |
ube3a
|
ubiquitin protein ligase E3A |
| chr20_-_20270191 | 0.43 |
ENSDART00000009356
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr4_+_15006217 | 0.43 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr17_+_33999630 | 0.43 |
ENSDART00000167085
ENSDART00000155030 ENSDART00000168522 ENSDART00000191799 ENSDART00000189684 ENSDART00000153942 ENSDART00000187272 ENSDART00000127692 |
gphna
|
gephyrin a |
| chr5_+_72145468 | 0.43 |
ENSDART00000148626
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr7_-_56766973 | 0.42 |
ENSDART00000020967
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr1_+_29183962 | 0.42 |
ENSDART00000113735
|
cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr10_+_20589969 | 0.42 |
ENSDART00000183042
|
letm2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr19_-_81477 | 0.42 |
ENSDART00000159815
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr23_-_4019699 | 0.42 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr23_-_45955177 | 0.42 |
ENSDART00000165963
ENSDART00000186649 ENSDART00000185773 |
LO017850.1
|
|
| chr14_-_16082806 | 0.42 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr12_-_17201028 | 0.41 |
ENSDART00000020541
|
lipf
|
lipase, gastric |
| chr2_+_27855346 | 0.41 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr18_+_15937610 | 0.41 |
ENSDART00000061134
ENSDART00000154505 |
itpr2
|
inositol 1,4,5-trisphosphate receptor, type 2 |
| chr8_-_31384607 | 0.41 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr13_+_13945218 | 0.41 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr13_-_42749916 | 0.41 |
ENSDART00000140019
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr20_+_47953047 | 0.41 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr2_-_41723487 | 0.41 |
ENSDART00000170171
|
zgc:110158
|
zgc:110158 |
| chr11_-_16021424 | 0.40 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr9_+_21536122 | 0.40 |
ENSDART00000182266
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr20_+_33924235 | 0.40 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr18_-_16907436 | 0.40 |
ENSDART00000146464
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr16_-_42770064 | 0.40 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr3_-_27880229 | 0.39 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr24_+_37484661 | 0.39 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr9_+_21535885 | 0.39 |
ENSDART00000141408
|
arhgef7a
|
Rho guanine nucleotide exchange factor (GEF) 7a |
| chr12_-_10505986 | 0.39 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr11_+_5880562 | 0.39 |
ENSDART00000129663
ENSDART00000130768 ENSDART00000160909 |
dazap1
|
DAZ associated protein 1 |
| chr5_-_37341044 | 0.39 |
ENSDART00000084675
|
wdr44
|
WD repeat domain 44 |
| chr3_-_50147160 | 0.39 |
ENSDART00000191341
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr14_-_689841 | 0.38 |
ENSDART00000125969
|
ube2ka
|
ubiquitin-conjugating enzyme E2Ka (UBC1 homolog, yeast) |
| chr13_+_35765317 | 0.38 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr6_+_40952031 | 0.38 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr7_-_59159253 | 0.38 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr1_+_24382651 | 0.37 |
ENSDART00000123789
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr5_-_22082918 | 0.37 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr7_+_16352924 | 0.37 |
ENSDART00000158972
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr2_+_30489846 | 0.37 |
ENSDART00000145732
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr8_-_17184482 | 0.37 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr11_+_26375979 | 0.37 |
ENSDART00000087652
ENSDART00000171748 ENSDART00000103513 ENSDART00000165931 ENSDART00000170043 |
cpne1
rbm12
|
copine I RNA binding motif protein 12 |
| chr21_-_38153824 | 0.37 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr20_+_32478151 | 0.36 |
ENSDART00000145269
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr22_+_1481307 | 0.36 |
ENSDART00000177429
|
si:ch211-255f4.2
|
si:ch211-255f4.2 |
| chr9_-_25366541 | 0.36 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr13_+_45524475 | 0.35 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr5_-_35200590 | 0.35 |
ENSDART00000051271
|
fcho2
|
FCH domain only 2 |
| chr23_+_44427188 | 0.35 |
ENSDART00000160207
|
eif4e2rs1
|
eukaryotic translation initiation factor 4E family member 2 related sequence 1 |
| chr6_+_4371137 | 0.35 |
ENSDART00000168973
|
rbm26
|
RNA binding motif protein 26 |
| chr8_+_23639124 | 0.35 |
ENSDART00000083108
|
nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr2_-_38117538 | 0.34 |
ENSDART00000013676
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr5_-_21030934 | 0.34 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr10_+_44955106 | 0.34 |
ENSDART00000185837
|
il1b
|
interleukin 1, beta |
| chr4_-_16124417 | 0.34 |
ENSDART00000128079
ENSDART00000077664 |
atp2b1a
|
ATPase plasma membrane Ca2+ transporting 1a |
| chr9_-_12575776 | 0.34 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr7_+_21768452 | 0.34 |
ENSDART00000127719
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr6_+_4370935 | 0.34 |
ENSDART00000192368
|
rbm26
|
RNA binding motif protein 26 |
| chr8_+_44623540 | 0.34 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr8_+_12951155 | 0.33 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr7_+_41812636 | 0.33 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr18_+_907266 | 0.33 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr18_-_49286381 | 0.33 |
ENSDART00000174248
ENSDART00000174038 |
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr5_-_3991655 | 0.33 |
ENSDART00000159368
|
myo19
|
myosin XIX |
| chr5_+_15992655 | 0.33 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr13_-_18011168 | 0.32 |
ENSDART00000144813
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr9_-_12575569 | 0.32 |
ENSDART00000102419
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr17_+_38030327 | 0.32 |
ENSDART00000085481
|
slc25a21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr3_+_17616201 | 0.32 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr20_-_28433616 | 0.32 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr17_+_47174525 | 0.32 |
ENSDART00000156831
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr1_-_47122058 | 0.32 |
ENSDART00000159925
ENSDART00000101143 ENSDART00000176803 |
mhc1zea
|
major histocompatibility complex class I ZEA |
| chr7_+_34794829 | 0.32 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis2a+meis2b_FO834857.1_pknox1.1+pknox1.2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.3 | 1.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 1.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.2 | GO:0070228 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.2 | 1.0 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 1.0 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.9 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.2 | 2.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 0.8 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.2 | 1.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.4 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.6 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.4 | GO:0060760 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.1 | 0.4 | GO:0021824 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.1 | 0.4 | GO:0045041 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.1 | 1.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.3 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.6 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 0.3 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 1.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.2 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.1 | 0.3 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 1.9 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.4 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 1.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.1 | 0.6 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 0.3 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.1 | 0.7 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.4 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.3 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.4 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.7 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.2 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 | 1.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.6 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 0.3 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.6 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 1.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.4 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 1.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 2.0 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 0.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0014015 | positive regulation of gliogenesis(GO:0014015) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.9 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 2.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.1 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.5 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.6 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 2.3 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 1.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.0 | GO:0070814 | cysteine biosynthetic process from serine(GO:0006535) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.7 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.5 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.0 | GO:0071042 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 1.0 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.7 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.6 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.1 | GO:0009437 | carnitine metabolic process(GO:0009437) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 1.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 1.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 0.6 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.1 | 0.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.3 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.3 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 1.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.7 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.6 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.9 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.4 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.0 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 1.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 1.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.4 | 1.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 2.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 0.6 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 0.8 | GO:0051800 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.2 | 1.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.2 | 1.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.4 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.7 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 1.2 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.4 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.4 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 0.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.9 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 1.0 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.7 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.6 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 1.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.6 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 3.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.8 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.3 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 2.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.2 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 4.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 2.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 1.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 1.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |