Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
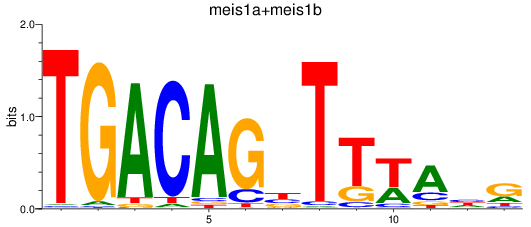
Results for meis1a+meis1b
Z-value: 0.68
Transcription factors associated with meis1a+meis1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
meis1a
|
ENSDARG00000002937 | Meis homeobox 1 a |
|
meis1b
|
ENSDARG00000012078 | Meis homeobox 1 b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| meis1b | dr11_v1_chr13_-_5569562_5569562 | -0.09 | 7.2e-01 | Click! |
| meis1a | dr11_v1_chr1_+_51475094_51475094 | 0.08 | 7.5e-01 | Click! |
Activity profile of meis1a+meis1b motif
Sorted Z-values of meis1a+meis1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_53092509 | 1.12 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr6_-_39199070 | 1.02 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr24_+_10027902 | 0.90 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr21_+_22330005 | 0.87 |
ENSDART00000140751
ENSDART00000157839 ENSDART00000140468 |
nadk2
|
NAD kinase 2, mitochondrial |
| chr24_-_10006158 | 0.80 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr5_+_35744623 | 0.78 |
ENSDART00000148213
ENSDART00000076627 |
yipf6
|
Yip1 domain family, member 6 |
| chr24_-_9985019 | 0.75 |
ENSDART00000193536
ENSDART00000189595 |
zgc:171977
|
zgc:171977 |
| chr4_-_18851365 | 0.75 |
ENSDART00000021782
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr8_-_17167819 | 0.74 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr11_-_544187 | 0.70 |
ENSDART00000173161
|
CU695207.1
|
|
| chr22_+_39084829 | 0.70 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr24_-_9979342 | 0.69 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr5_-_2721686 | 0.64 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr24_-_10014512 | 0.62 |
ENSDART00000124341
ENSDART00000191630 |
zgc:171474
|
zgc:171474 |
| chr12_+_46543572 | 0.61 |
ENSDART00000167510
|
hid1b
|
HID1 domain containing b |
| chr2_+_7557912 | 0.58 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr6_-_7052595 | 0.57 |
ENSDART00000081761
ENSDART00000181351 |
bin1b
|
bridging integrator 1b |
| chr7_-_16034324 | 0.52 |
ENSDART00000002498
ENSDART00000162962 |
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr7_-_19369002 | 0.51 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr19_+_20778011 | 0.51 |
ENSDART00000024208
|
nutf2l
|
nuclear transport factor 2, like |
| chr13_-_8692860 | 0.50 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr2_+_27855346 | 0.50 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr10_-_26512742 | 0.49 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr9_+_38399912 | 0.49 |
ENSDART00000022246
ENSDART00000145892 |
bcs1l
|
BC1 (ubiquinol-cytochrome c reductase) synthesis-like |
| chr20_-_30035326 | 0.49 |
ENSDART00000141068
|
sox11b
|
SRY (sex determining region Y)-box 11b |
| chr13_-_8692432 | 0.49 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr17_+_33375469 | 0.47 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr11_+_1796426 | 0.47 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr6_-_7052408 | 0.47 |
ENSDART00000150033
ENSDART00000149232 |
bin1b
|
bridging integrator 1b |
| chr4_-_5332814 | 0.46 |
ENSDART00000144225
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr4_-_18850799 | 0.46 |
ENSDART00000151844
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr21_+_17024002 | 0.45 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr4_-_15603511 | 0.45 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr3_-_20793655 | 0.44 |
ENSDART00000163473
ENSDART00000159457 |
spop
|
speckle-type POZ protein |
| chr14_-_24251057 | 0.43 |
ENSDART00000114169
|
bnip1a
|
BCL2 interacting protein 1a |
| chr25_-_37314700 | 0.43 |
ENSDART00000017807
|
igl4v8
|
immunoglobulin light 4 variable 8 |
| chr21_-_2958422 | 0.43 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr1_-_53880639 | 0.42 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr13_+_33246150 | 0.42 |
ENSDART00000136356
ENSDART00000132834 ENSDART00000144379 ENSDART00000131985 ENSDART00000148368 ENSDART00000035940 ENSDART00000141912 |
ndufaf1
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr22_-_38034852 | 0.42 |
ENSDART00000104613
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr10_-_45029041 | 0.42 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr7_-_18881358 | 0.41 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr8_-_45838277 | 0.41 |
ENSDART00000046064
|
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr22_-_38035084 | 0.41 |
ENSDART00000145029
|
pex6
|
peroxisomal biogenesis factor 6 |
| chr3_+_17537352 | 0.41 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr7_+_55292959 | 0.40 |
ENSDART00000147539
ENSDART00000073555 |
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr12_+_4220353 | 0.40 |
ENSDART00000133675
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr8_+_53051701 | 0.40 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr8_-_45838481 | 0.40 |
ENSDART00000148206
ENSDART00000170485 |
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr15_-_24588649 | 0.38 |
ENSDART00000060469
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr24_-_2736809 | 0.38 |
ENSDART00000171805
|
fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr15_-_684911 | 0.37 |
ENSDART00000161685
|
zgc:171901
|
zgc:171901 |
| chr10_-_42776344 | 0.37 |
ENSDART00000190653
ENSDART00000130229 |
wdr45
|
WD repeat domain 45 |
| chr7_-_7676186 | 0.36 |
ENSDART00000091105
|
mfap3l
|
microfibril associated protein 3 like |
| chr20_-_37813863 | 0.36 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr10_+_24692076 | 0.35 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr16_+_4695075 | 0.34 |
ENSDART00000039054
|
mecr
|
mitochondrial trans-2-enoyl-CoA reductase |
| chr20_-_26391958 | 0.34 |
ENSDART00000078062
|
armt1
|
acidic residue methyltransferase 1 |
| chr5_-_21030934 | 0.34 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr13_+_24584401 | 0.34 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr17_+_24809221 | 0.33 |
ENSDART00000082251
ENSDART00000147871 ENSDART00000130871 |
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr14_+_2243 | 0.33 |
ENSDART00000191193
|
CYTL1
|
cytokine like 1 |
| chr14_-_28473639 | 0.33 |
ENSDART00000173007
|
nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr20_+_3339248 | 0.31 |
ENSDART00000108955
|
mfsd4b
|
major facilitator superfamily domain containing 4B |
| chr5_-_54497319 | 0.31 |
ENSDART00000160492
|
alad
|
aminolevulinate dehydratase |
| chr10_-_26512993 | 0.30 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr24_-_26369185 | 0.30 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr4_-_14191717 | 0.30 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr3_-_1400309 | 0.30 |
ENSDART00000159893
|
wbp11
|
WW domain binding protein 11 |
| chr5_+_55984270 | 0.30 |
ENSDART00000047358
ENSDART00000138191 |
fkbp6
|
FK506 binding protein 6 |
| chr13_-_7031033 | 0.29 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr4_-_14192254 | 0.29 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr12_-_31461219 | 0.29 |
ENSDART00000148954
|
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr7_-_19526721 | 0.29 |
ENSDART00000114203
|
man2b2
|
mannosidase, alpha, class 2B, member 2 |
| chr2_+_51645164 | 0.28 |
ENSDART00000169600
|
abhd4
|
abhydrolase domain containing 4 |
| chr10_-_43117607 | 0.28 |
ENSDART00000148293
ENSDART00000089965 |
tmem167a
|
transmembrane protein 167A |
| chr4_-_14191434 | 0.28 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr15_-_25365319 | 0.27 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr4_+_16768961 | 0.27 |
ENSDART00000143926
|
BX649500.1
|
|
| chr5_-_66215928 | 0.27 |
ENSDART00000082797
|
coq2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr20_-_7176809 | 0.26 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr4_+_16777715 | 0.26 |
ENSDART00000148344
|
BX649476.1
|
|
| chr2_-_41861040 | 0.26 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr6_-_16493516 | 0.26 |
ENSDART00000022499
|
psmb3
|
proteasome subunit beta 3 |
| chr5_-_67750907 | 0.25 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr3_-_7134113 | 0.25 |
ENSDART00000180849
|
BX005085.6
|
|
| chr8_-_31384607 | 0.25 |
ENSDART00000164134
ENSDART00000024872 |
creb3l3l
|
cAMP responsive element binding protein 3-like 3 like |
| chr8_-_13972626 | 0.25 |
ENSDART00000144296
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr2_+_19236969 | 0.24 |
ENSDART00000163875
ENSDART00000168644 |
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr2_-_44971551 | 0.24 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr19_+_619200 | 0.24 |
ENSDART00000050125
|
nupl2
|
nucleoporin like 2 |
| chr19_+_33850705 | 0.24 |
ENSDART00000160356
|
pex1
|
peroxisomal biogenesis factor 1 |
| chr1_+_58628165 | 0.24 |
ENSDART00000158654
|
si:ch73-221f6.4
|
si:ch73-221f6.4 |
| chr14_+_22172047 | 0.23 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr23_+_40410644 | 0.23 |
ENSDART00000056328
|
elovl4b
|
ELOVL fatty acid elongase 4b |
| chr25_+_37297659 | 0.23 |
ENSDART00000086733
|
si:dkey-234i14.13
|
si:dkey-234i14.13 |
| chr23_+_23183449 | 0.22 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr6_+_54154807 | 0.22 |
ENSDART00000061735
|
nudt3b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3b |
| chr20_+_26987416 | 0.22 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr23_+_23182037 | 0.22 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr13_+_24287093 | 0.22 |
ENSDART00000058628
|
ccsapb
|
centriole, cilia and spindle-associated protein b |
| chr9_+_33222911 | 0.22 |
ENSDART00000135387
|
si:ch211-125e6.14
|
si:ch211-125e6.14 |
| chr16_-_30826712 | 0.22 |
ENSDART00000122474
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr14_-_5546497 | 0.21 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr2_-_26469065 | 0.21 |
ENSDART00000099247
ENSDART00000099248 |
rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr21_+_11749097 | 0.21 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr20_+_38910214 | 0.21 |
ENSDART00000047362
|
msra
|
methionine sulfoxide reductase A |
| chr19_+_7453996 | 0.21 |
ENSDART00000147491
ENSDART00000136502 |
mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr12_+_34273240 | 0.20 |
ENSDART00000037904
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr3_-_55104310 | 0.20 |
ENSDART00000101713
|
hbba1
|
hemoglobin, beta adult 1 |
| chr19_+_48111285 | 0.19 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr18_-_19006634 | 0.19 |
ENSDART00000060731
|
ints14
|
integrator complex subunit 14 |
| chr4_+_7876197 | 0.19 |
ENSDART00000111986
ENSDART00000189601 |
cdc123
|
cell division cycle 123 homolog (S. cerevisiae) |
| chr7_-_41013575 | 0.19 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr2_-_22993601 | 0.19 |
ENSDART00000125049
ENSDART00000099735 |
mllt1b
|
MLLT1, super elongation complex subunit b |
| chr3_+_25990052 | 0.19 |
ENSDART00000007398
|
gcat
|
glycine C-acetyltransferase |
| chr5_+_37744625 | 0.18 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr1_+_58614897 | 0.18 |
ENSDART00000167354
ENSDART00000162849 |
si:ch73-221f6.3
|
si:ch73-221f6.3 |
| chr6_+_49926115 | 0.18 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr1_-_19648227 | 0.18 |
ENSDART00000054574
|
polr1e
|
polymerase (RNA) I polypeptide E |
| chr10_-_45210184 | 0.18 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr23_-_10254288 | 0.18 |
ENSDART00000081215
|
krt8
|
keratin 8 |
| chr14_-_26425416 | 0.18 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr10_+_39476600 | 0.18 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr2_-_8609653 | 0.18 |
ENSDART00000193354
ENSDART00000189489 ENSDART00000186144 |
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr12_+_11167870 | 0.17 |
ENSDART00000052537
|
vps35l
|
zgc:163107 |
| chr17_-_43594864 | 0.17 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr3_+_29641181 | 0.17 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr17_-_30839338 | 0.17 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr5_-_26284276 | 0.17 |
ENSDART00000148608
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr22_-_24791505 | 0.17 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr10_-_24765988 | 0.16 |
ENSDART00000064463
|
timm10b
|
translocase of inner mitochondrial membrane 10 homolog B (yeast) |
| chr25_+_19670273 | 0.16 |
ENSDART00000073472
|
zgc:113426
|
zgc:113426 |
| chr8_-_22508055 | 0.16 |
ENSDART00000101616
|
si:ch211-261n11.5
|
si:ch211-261n11.5 |
| chr11_+_41540862 | 0.16 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr5_-_18446483 | 0.16 |
ENSDART00000180027
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr7_-_55292116 | 0.16 |
ENSDART00000122603
|
rnf166
|
ring finger protein 166 |
| chr25_+_34641536 | 0.16 |
ENSDART00000167033
|
CABZ01079011.1
|
|
| chr5_-_33298352 | 0.16 |
ENSDART00000015076
|
zgc:103692
|
zgc:103692 |
| chr22_+_35275468 | 0.15 |
ENSDART00000189516
ENSDART00000181572 ENSDART00000165353 ENSDART00000185352 |
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr15_+_24588963 | 0.15 |
ENSDART00000155075
|
zgc:198241
|
zgc:198241 |
| chr10_-_39011514 | 0.15 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr11_+_27347076 | 0.14 |
ENSDART00000173383
|
fbln2
|
fibulin 2 |
| chr4_-_77561679 | 0.14 |
ENSDART00000180809
|
AL935186.9
|
|
| chr21_-_26483237 | 0.14 |
ENSDART00000169072
ENSDART00000147947 |
ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr6_+_9107063 | 0.14 |
ENSDART00000083820
|
vps16
|
vacuolar protein sorting protein 16 |
| chr8_+_41413919 | 0.14 |
ENSDART00000142590
|
si:ch211-152p23.2
|
si:ch211-152p23.2 |
| chr9_+_38168012 | 0.14 |
ENSDART00000102445
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr5_+_50898849 | 0.14 |
ENSDART00000083317
|
arsk
|
arylsulfatase family, member K |
| chr10_+_29770120 | 0.14 |
ENSDART00000100032
ENSDART00000193205 |
hyou1
|
hypoxia up-regulated 1 |
| chr22_+_1006573 | 0.14 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr1_+_39859782 | 0.14 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr5_+_51248784 | 0.14 |
ENSDART00000159571
ENSDART00000189513 ENSDART00000092285 |
msh3
|
mutS homolog 3 (E. coli) |
| chr25_-_17852944 | 0.14 |
ENSDART00000155169
|
si:ch211-176l24.4
|
si:ch211-176l24.4 |
| chr16_-_33817828 | 0.14 |
ENSDART00000188395
|
rspo1
|
R-spondin 1 |
| chr25_-_3192405 | 0.14 |
ENSDART00000104835
|
hps5
|
Hermansky-Pudlak syndrome 5 |
| chr5_+_56119975 | 0.14 |
ENSDART00000083137
|
mrm1
|
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
| chr4_-_20208166 | 0.14 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr1_+_35473397 | 0.13 |
ENSDART00000158799
|
usp38
|
ubiquitin specific peptidase 38 |
| chr1_+_58609885 | 0.13 |
ENSDART00000186199
|
si:ch73-236c18.9
|
si:ch73-236c18.9 |
| chr11_-_18557277 | 0.13 |
ENSDART00000103950
|
dido1
|
death inducer-obliterator 1 |
| chr1_+_34496855 | 0.13 |
ENSDART00000012873
|
klf12a
|
Kruppel-like factor 12a |
| chr23_+_22335407 | 0.13 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr1_-_49919042 | 0.13 |
ENSDART00000050603
|
hadh
|
hydroxyacyl-CoA dehydrogenase |
| chr8_+_41453691 | 0.13 |
ENSDART00000061144
|
arpc5la
|
actin related protein 2/3 complex, subunit 5-like, a |
| chr2_+_11028923 | 0.13 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr22_+_35275206 | 0.13 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr24_+_2736815 | 0.13 |
ENSDART00000184009
|
LYRM4
|
si:ch211-152c8.4 |
| chr2_+_24374669 | 0.13 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr15_-_9294929 | 0.13 |
ENSDART00000155717
|
si:ch211-261a10.5
|
si:ch211-261a10.5 |
| chr7_-_17814118 | 0.13 |
ENSDART00000179688
|
ecsit
|
ECSIT signalling integrator |
| chr24_+_9881219 | 0.13 |
ENSDART00000036204
|
cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr7_+_40205394 | 0.13 |
ENSDART00000173742
ENSDART00000148390 |
ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr24_-_13272842 | 0.12 |
ENSDART00000146790
ENSDART00000066703 |
rdh10a
|
retinol dehydrogenase 10a |
| chr17_+_39741926 | 0.12 |
ENSDART00000154996
ENSDART00000154599 |
si:dkey-229e3.2
|
si:dkey-229e3.2 |
| chr6_+_33881372 | 0.12 |
ENSDART00000165710
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr15_+_41027466 | 0.12 |
ENSDART00000075940
|
mtnr1ba
|
melatonin receptor type 1Ba |
| chr5_+_26079178 | 0.12 |
ENSDART00000145920
|
si:dkey-201c13.2
|
si:dkey-201c13.2 |
| chr20_+_23855025 | 0.12 |
ENSDART00000032393
|
ginm1
|
glycoprotein integral membrane 1 |
| chr14_+_29941266 | 0.12 |
ENSDART00000112757
|
fam149a
|
family with sequence similarity 149 member A |
| chr3_-_40162843 | 0.12 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr20_-_44460789 | 0.12 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr21_-_28523548 | 0.12 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr13_+_40692804 | 0.12 |
ENSDART00000109822
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr22_+_1276680 | 0.12 |
ENSDART00000169230
|
si:ch73-138e16.2
|
si:ch73-138e16.2 |
| chr21_+_37357578 | 0.12 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr4_-_15420452 | 0.12 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr3_-_55099272 | 0.12 |
ENSDART00000130869
|
hbba1
|
hemoglobin, beta adult 1 |
| chr11_-_21303946 | 0.11 |
ENSDART00000185786
|
RASSF5
|
si:dkey-85p17.3 |
| chr12_+_48841419 | 0.11 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr12_+_18768021 | 0.11 |
ENSDART00000153111
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr20_-_5267600 | 0.11 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr23_-_4930229 | 0.11 |
ENSDART00000189456
|
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr23_-_31555696 | 0.11 |
ENSDART00000053539
|
tcf21
|
transcription factor 21 |
| chr11_-_13151841 | 0.11 |
ENSDART00000161983
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr2_+_6127593 | 0.11 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr11_+_31558207 | 0.11 |
ENSDART00000140204
|
egln1b
|
egl-9 family hypoxia-inducible factor 1b |
| chr19_+_10855158 | 0.11 |
ENSDART00000172219
ENSDART00000170826 |
apoea
|
apolipoprotein Ea |
| chr4_-_20235904 | 0.11 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr8_-_25605537 | 0.11 |
ENSDART00000005906
|
stk38a
|
serine/threonine kinase 38a |
Network of associatons between targets according to the STRING database.
First level regulatory network of meis1a+meis1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.5 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 0.3 | GO:2000193 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.2 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.2 | GO:0036316 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 1.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.1 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.2 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.5 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.6 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.0 | 0.1 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.4 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.0 | 0.1 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.1 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.0 | 0.5 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.6 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.3 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0042144 | vacuole fusion, non-autophagic(GO:0042144) |
| 0.0 | 0.7 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.1 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 1.5 | GO:0006633 | fatty acid biosynthetic process(GO:0006633) |
| 0.0 | 0.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.0 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.0 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.0 | 0.8 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.1 | GO:0010517 | regulation of phospholipase activity(GO:0010517) lipid digestion(GO:0044241) |
| 0.0 | 0.8 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.3 | 0.8 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 0.7 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.9 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.2 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.5 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.2 | 1.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.2 | 0.5 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 0.8 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.3 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.6 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 1.5 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.2 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 1.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.4 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.1 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.3 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.6 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |