Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
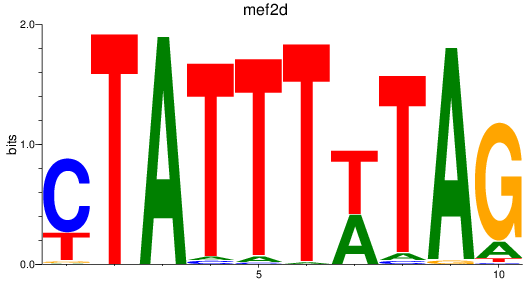
Results for mef2d
Z-value: 0.82
Transcription factors associated with mef2d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2d
|
ENSDARG00000040237 | myocyte enhancer factor 2d |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2d | dr11_v1_chr16_-_29164379_29164379 | 0.74 | 4.2e-04 | Click! |
Activity profile of mef2d motif
Sorted Z-values of mef2d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_36948586 | 2.26 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr3_+_28953274 | 2.11 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr22_-_906757 | 2.05 |
ENSDART00000193182
|
FP016205.1
|
|
| chr10_-_22845485 | 1.89 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr5_-_38643188 | 1.89 |
ENSDART00000144972
|
si:ch211-271e10.1
|
si:ch211-271e10.1 |
| chr16_+_34523515 | 1.74 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr17_-_5769196 | 1.70 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr10_-_29816467 | 1.51 |
ENSDART00000055913
|
hist2h2l
|
histone 2, H2, like |
| chr20_-_22778394 | 1.49 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr12_-_36521767 | 1.46 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr15_+_40008370 | 1.46 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr7_-_28148310 | 1.45 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr8_-_50259448 | 1.43 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr8_+_44703864 | 1.36 |
ENSDART00000016225
|
star
|
steroidogenic acute regulatory protein |
| chr5_-_72125551 | 1.27 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr2_+_49569017 | 1.21 |
ENSDART00000109471
|
sema4e
|
semaphorin 4e |
| chr23_-_23401305 | 1.18 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr25_-_4482449 | 1.17 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr10_+_29816681 | 1.17 |
ENSDART00000100022
|
h2afx1
|
H2A histone family member X1 |
| chr4_-_12795030 | 1.15 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr20_+_41549200 | 1.15 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr23_-_40766518 | 1.11 |
ENSDART00000127420
|
si:dkeyp-27c8.2
|
si:dkeyp-27c8.2 |
| chr8_+_22930627 | 1.10 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr5_-_63286077 | 1.10 |
ENSDART00000131274
|
tbcelb
|
tubulin folding cofactor E-like b |
| chr21_+_40589770 | 1.10 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr24_-_4765740 | 1.08 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr4_-_9592402 | 1.04 |
ENSDART00000114060
|
cdnf
|
cerebral dopamine neurotrophic factor |
| chr2_+_6181383 | 1.04 |
ENSDART00000153307
|
si:ch73-344o19.1
|
si:ch73-344o19.1 |
| chr14_+_23520986 | 1.04 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr3_-_52674089 | 1.03 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr5_+_62723233 | 1.02 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr10_-_3332362 | 1.01 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr10_-_24371312 | 1.01 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr5_-_19394440 | 0.99 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr25_-_16554757 | 0.99 |
ENSDART00000154480
|
si:ch211-266k8.6
|
si:ch211-266k8.6 |
| chr13_-_37647209 | 0.99 |
ENSDART00000189102
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr17_-_10025234 | 0.99 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr22_+_16320076 | 0.96 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr4_-_12795436 | 0.95 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr14_+_24215046 | 0.93 |
ENSDART00000079215
|
stc2a
|
stanniocalcin 2a |
| chr5_-_10946232 | 0.92 |
ENSDART00000163139
ENSDART00000031265 |
rtn4r
|
reticulon 4 receptor |
| chr13_-_28674422 | 0.89 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr1_+_7546259 | 0.88 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr16_-_31919568 | 0.87 |
ENSDART00000027364
|
rbfox1l
|
RNA binding fox-1 homolog 1, like |
| chr25_-_7753207 | 0.86 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr23_+_20110086 | 0.86 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr3_-_31655796 | 0.85 |
ENSDART00000181052
|
CU062431.1
|
|
| chr14_-_36799280 | 0.83 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr25_+_32530976 | 0.82 |
ENSDART00000156190
ENSDART00000103324 |
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr23_+_20687340 | 0.81 |
ENSDART00000143503
|
usp21
|
ubiquitin specific peptidase 21 |
| chr9_-_21382190 | 0.81 |
ENSDART00000110943
ENSDART00000188129 ENSDART00000193876 |
ift88
|
intraflagellar transport 88 homolog |
| chr21_+_22828500 | 0.80 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr15_+_37412883 | 0.79 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr22_+_15898221 | 0.78 |
ENSDART00000062587
|
klf2a
|
Kruppel-like factor 2a |
| chr1_-_58592866 | 0.78 |
ENSDART00000161039
|
adgre5b.2
|
adhesion G protein-coupled receptor E5b, duplicate 2 |
| chr22_+_16497670 | 0.77 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr23_+_32011768 | 0.77 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr24_+_17068724 | 0.77 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr8_+_17987215 | 0.77 |
ENSDART00000113605
|
lrriq3
|
leucine-rich repeats and IQ motif containing 3 |
| chr17_+_42842632 | 0.76 |
ENSDART00000155547
ENSDART00000126087 |
qpct
|
glutaminyl-peptide cyclotransferase |
| chr23_+_20705849 | 0.76 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr11_+_11201096 | 0.76 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr24_+_17069420 | 0.76 |
ENSDART00000014787
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr21_-_5799122 | 0.74 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr9_+_31795343 | 0.74 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr7_+_14291323 | 0.73 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr9_-_23891102 | 0.72 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr22_+_38310957 | 0.72 |
ENSDART00000040550
|
traf5
|
Tnf receptor-associated factor 5 |
| chr6_-_40722480 | 0.71 |
ENSDART00000188187
|
kbtbd12
|
kelch repeat and BTB (POZ) domain containing 12 |
| chr12_-_17707449 | 0.71 |
ENSDART00000142427
ENSDART00000034914 |
pvalb3
|
parvalbumin 3 |
| chr3_+_32425202 | 0.70 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr9_-_43142636 | 0.70 |
ENSDART00000134349
ENSDART00000181835 |
ccdc141
|
coiled-coil domain containing 141 |
| chr12_-_19007834 | 0.70 |
ENSDART00000153248
|
chadlb
|
chondroadherin-like b |
| chr22_-_31059670 | 0.69 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr23_+_44497701 | 0.68 |
ENSDART00000149903
|
si:ch73-375g18.1
|
si:ch73-375g18.1 |
| chr3_+_18398876 | 0.66 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr8_-_46321889 | 0.66 |
ENSDART00000075189
ENSDART00000122801 |
mtor
|
mechanistic target of rapamycin kinase |
| chr19_+_40983221 | 0.64 |
ENSDART00000144544
|
col1a2
|
collagen, type I, alpha 2 |
| chr25_+_13791627 | 0.64 |
ENSDART00000159278
|
zgc:92873
|
zgc:92873 |
| chr7_-_40993456 | 0.64 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr3_+_18399315 | 0.64 |
ENSDART00000110842
|
rps2
|
ribosomal protein S2 |
| chr17_+_32374876 | 0.63 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr20_+_46371458 | 0.63 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr21_+_28445052 | 0.63 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr17_-_13026634 | 0.62 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr1_-_11075403 | 0.61 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr23_+_23022790 | 0.61 |
ENSDART00000189843
|
samd11
|
sterile alpha motif domain containing 11 |
| chr8_-_25002182 | 0.61 |
ENSDART00000078792
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr16_+_16968682 | 0.60 |
ENSDART00000111074
|
si:ch211-120k19.1
|
si:ch211-120k19.1 |
| chr2_-_43168292 | 0.58 |
ENSDART00000132588
|
crema
|
cAMP responsive element modulator a |
| chr10_-_22803740 | 0.58 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr24_-_24038800 | 0.58 |
ENSDART00000080549
|
lyz
|
lysozyme |
| chr9_-_34300707 | 0.58 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr17_+_7513673 | 0.58 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr16_-_22713152 | 0.58 |
ENSDART00000140953
ENSDART00000143836 |
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr3_+_18555899 | 0.58 |
ENSDART00000163036
ENSDART00000158791 |
cbx4
|
chromobox homolog 4 (Pc class homolog, Drosophila) |
| chr2_+_2737422 | 0.57 |
ENSDART00000032459
|
aqp1a.1
|
aquaporin 1a (Colton blood group), tandem duplicate 1 |
| chr21_-_22828593 | 0.56 |
ENSDART00000150993
|
angptl5
|
angiopoietin-like 5 |
| chr16_+_16969060 | 0.56 |
ENSDART00000182819
ENSDART00000191876 |
si:ch211-120k19.1
rpl18
|
si:ch211-120k19.1 ribosomal protein L18 |
| chr14_-_30704075 | 0.56 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr9_+_17438765 | 0.56 |
ENSDART00000138953
|
rgcc
|
regulator of cell cycle |
| chr1_-_411331 | 0.55 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr10_+_2981090 | 0.55 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr5_-_48268049 | 0.55 |
ENSDART00000187454
|
mef2cb
|
myocyte enhancer factor 2cb |
| chr20_+_812012 | 0.55 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr2_-_16562505 | 0.55 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr21_+_19070921 | 0.55 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr20_+_15552657 | 0.54 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr4_+_11375894 | 0.54 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr20_+_16881883 | 0.53 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr25_+_3677650 | 0.53 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr1_-_53907092 | 0.53 |
ENSDART00000007732
|
capn9
|
calpain 9 |
| chr19_-_38611814 | 0.52 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr12_+_6002715 | 0.51 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr10_-_4980150 | 0.51 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr20_-_36617313 | 0.50 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr2_+_29976419 | 0.50 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr8_-_25728628 | 0.49 |
ENSDART00000127237
|
foxp3a
|
forkhead box P3a |
| chr4_-_57530817 | 0.49 |
ENSDART00000158435
|
zgc:173702
|
zgc:173702 |
| chr8_-_26388090 | 0.49 |
ENSDART00000147912
|
si:dkey-20d21.12
|
si:dkey-20d21.12 |
| chr25_-_3644574 | 0.49 |
ENSDART00000188569
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr17_+_15899039 | 0.48 |
ENSDART00000154181
|
syne3
|
spectrin repeat containing, nuclear envelope family member 3 |
| chr8_+_52637507 | 0.48 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr14_-_21064199 | 0.48 |
ENSDART00000172099
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr14_-_21063977 | 0.47 |
ENSDART00000164373
|
si:dkey-74k8.3
|
si:dkey-74k8.3 |
| chr16_-_24518027 | 0.47 |
ENSDART00000134120
ENSDART00000143761 |
cadm4
|
cell adhesion molecule 4 |
| chr10_+_29698467 | 0.46 |
ENSDART00000163402
|
dlg2
|
discs, large homolog 2 (Drosophila) |
| chr5_-_22619879 | 0.46 |
ENSDART00000051623
|
zgc:113208
|
zgc:113208 |
| chr12_+_33894665 | 0.45 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr11_-_3860534 | 0.45 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr15_+_47903864 | 0.45 |
ENSDART00000063835
|
otx5
|
orthodenticle homolog 5 |
| chr9_-_52814204 | 0.45 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr9_-_12424791 | 0.45 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr7_-_7420301 | 0.44 |
ENSDART00000102620
|
six7
|
SIX homeobox 7 |
| chr11_-_18601955 | 0.44 |
ENSDART00000180565
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr22_-_29204960 | 0.44 |
ENSDART00000131386
|
pvalb7
|
parvalbumin 7 |
| chr15_-_23645810 | 0.44 |
ENSDART00000168845
|
ckmb
|
creatine kinase, muscle b |
| chr23_+_6077503 | 0.44 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr18_+_30567945 | 0.43 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr12_-_36521947 | 0.43 |
ENSDART00000152946
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr7_+_31879649 | 0.43 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr1_-_6494384 | 0.42 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr14_-_30366196 | 0.42 |
ENSDART00000007022
|
pdgfrl
|
platelet-derived growth factor receptor-like |
| chr19_-_25464291 | 0.42 |
ENSDART00000112915
|
umad1
|
UBAP1-MVB12-associated (UMA) domain containing 1 |
| chr7_-_18547420 | 0.41 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr6_+_29410986 | 0.41 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 |
| chr18_-_2639351 | 0.41 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr14_-_24081929 | 0.41 |
ENSDART00000158576
|
msx2a
|
muscle segment homeobox 2a |
| chr21_-_22951604 | 0.41 |
ENSDART00000083449
ENSDART00000180129 |
dub
|
duboraya |
| chr12_+_16440708 | 0.40 |
ENSDART00000113810
|
ankrd1b
|
ankyrin repeat domain 1b (cardiac muscle) |
| chr13_-_37636147 | 0.40 |
ENSDART00000144065
|
si:dkey-188i13.8
|
si:dkey-188i13.8 |
| chr8_+_17184602 | 0.40 |
ENSDART00000050228
ENSDART00000140531 |
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr19_+_3056450 | 0.40 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr24_-_35534273 | 0.39 |
ENSDART00000026578
|
ube2v2
|
ubiquitin-conjugating enzyme E2 variant 2 |
| chr7_+_38811800 | 0.39 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr5_+_66132394 | 0.39 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr15_+_15856178 | 0.39 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr22_+_10606863 | 0.38 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr25_+_31264155 | 0.38 |
ENSDART00000012256
|
tnni2a.3
|
troponin I type 2a (skeletal, fast), tandem duplicate 3 |
| chr17_-_24439672 | 0.38 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr16_-_32975951 | 0.37 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr16_+_29663809 | 0.37 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr2_-_22492289 | 0.37 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr22_+_29204885 | 0.37 |
ENSDART00000133409
ENSDART00000059820 |
ncf4
|
neutrophil cytosolic factor 4 |
| chr23_+_23485858 | 0.36 |
ENSDART00000114067
|
agrn
|
agrin |
| chr3_-_14103551 | 0.36 |
ENSDART00000127604
|
nfil3-6
|
nuclear factor, interleukin 3 regulated, member 6 |
| chr11_+_40148181 | 0.36 |
ENSDART00000125119
|
si:dkey-264d12.5
|
si:dkey-264d12.5 |
| chr2_-_6182098 | 0.35 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr25_+_20716176 | 0.35 |
ENSDART00000073651
|
ergic2
|
ERGIC and golgi 2 |
| chr22_+_10606573 | 0.35 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr13_-_11536951 | 0.35 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr21_+_27448856 | 0.35 |
ENSDART00000100784
|
cfbl
|
complement factor b-like |
| chr18_+_17959681 | 0.34 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr25_+_29160102 | 0.34 |
ENSDART00000162854
|
pkmb
|
pyruvate kinase M1/2b |
| chr16_+_5259886 | 0.34 |
ENSDART00000186668
|
plecb
|
plectin b |
| chr11_-_39202915 | 0.34 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr3_-_13946446 | 0.33 |
ENSDART00000171249
|
gcdhb
|
glutaryl-CoA dehydrogenase b |
| chr3_+_52442102 | 0.33 |
ENSDART00000155455
|
si:ch211-191c10.1
|
si:ch211-191c10.1 |
| chr1_+_8601935 | 0.33 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr8_+_23147218 | 0.33 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr25_+_14092871 | 0.33 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr2_+_9552456 | 0.33 |
ENSDART00000056896
|
dnajb4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr17_-_12336987 | 0.32 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr23_-_18030399 | 0.32 |
ENSDART00000136967
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr14_+_1240419 | 0.32 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr6_-_40697585 | 0.31 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr20_-_38836161 | 0.31 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr5_+_28041967 | 0.31 |
ENSDART00000133641
|
ogfod2
|
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
| chr1_-_44638058 | 0.31 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr9_+_44430974 | 0.31 |
ENSDART00000056846
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr6_+_54187643 | 0.31 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr11_-_3954691 | 0.30 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr11_+_11200550 | 0.30 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr4_-_20313810 | 0.30 |
ENSDART00000136350
|
dcp1b
|
decapping mRNA 1B |
| chr11_-_29685853 | 0.29 |
ENSDART00000128468
|
nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr15_+_19885498 | 0.29 |
ENSDART00000152123
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr13_-_32635859 | 0.29 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr20_-_39271844 | 0.29 |
ENSDART00000192708
|
clu
|
clusterin |
| chr13_-_50139916 | 0.28 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr15_+_19652807 | 0.28 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.3 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 0.8 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.3 | 0.8 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 0.2 | 1.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 1.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.6 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 0.8 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.9 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 0.6 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) |
| 0.2 | 0.5 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 1.5 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 1.0 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.2 | 2.1 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.2 | 0.5 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 1.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 1.9 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.4 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 1.3 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.5 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 1.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 0.4 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.5 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.9 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 0.2 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.1 | 0.4 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.5 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.1 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.3 | GO:0008584 | male gonad development(GO:0008584) water homeostasis(GO:0030104) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 1.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 2.0 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.9 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.8 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.5 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 1.5 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.0 | 0.6 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.3 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.5 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.2 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.0 | 0.1 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.7 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.9 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.0 | 0.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.5 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0045197 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 1.6 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.0 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.7 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.4 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.3 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.5 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.9 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.5 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.6 | GO:0060914 | heart formation(GO:0060914) |
| 0.0 | 0.5 | GO:0030901 | midbrain development(GO:0030901) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 2.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.2 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 0.7 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 1.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.5 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.6 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 1.0 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.2 | 2.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.6 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 0.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.6 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.5 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.6 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 1.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 1.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.3 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 3.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 0.3 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 0.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.7 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.4 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.9 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 2.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.3 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.0 | 0.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 2.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.4 | 2.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.8 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.9 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |