Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
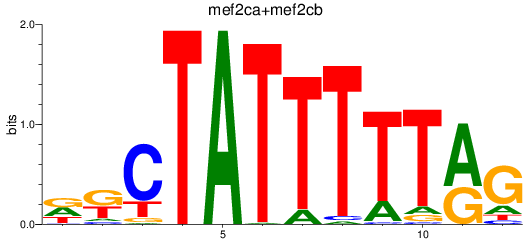
Results for mef2ca+mef2cb
Z-value: 0.75
Transcription factors associated with mef2ca+mef2cb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2cb
|
ENSDARG00000009418 | myocyte enhancer factor 2cb |
|
mef2ca
|
ENSDARG00000029764 | myocyte enhancer factor 2ca |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2cb | dr11_v1_chr5_-_48307804_48307804 | -0.75 | 3.8e-04 | Click! |
| mef2ca | dr11_v1_chr10_-_43568239_43568241 | -0.69 | 1.5e-03 | Click! |
Activity profile of mef2ca+mef2cb motif
Sorted Z-values of mef2ca+mef2cb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_49031303 | 2.80 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr13_-_21672131 | 2.12 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr21_+_25777425 | 2.12 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr4_-_1720648 | 2.08 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr7_+_38349667 | 2.00 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr1_+_12302073 | 1.95 |
ENSDART00000164045
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr2_-_43545342 | 1.83 |
ENSDART00000179796
|
CABZ01058721.1
|
|
| chr14_+_30291611 | 1.79 |
ENSDART00000173004
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr14_-_33481428 | 1.79 |
ENSDART00000147059
ENSDART00000140001 ENSDART00000124242 ENSDART00000164836 ENSDART00000190104 ENSDART00000186833 ENSDART00000180873 |
lamp2
|
lysosomal-associated membrane protein 2 |
| chr20_-_487783 | 1.74 |
ENSDART00000162218
|
col10a1b
|
collagen, type X, alpha 1b |
| chr11_-_34577034 | 1.70 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr6_+_3717613 | 1.68 |
ENSDART00000184330
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr6_-_49547680 | 1.63 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr6_+_38896158 | 1.62 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr9_+_21793565 | 1.61 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr14_-_7245971 | 1.61 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr21_-_19919918 | 1.57 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr22_-_22164338 | 1.55 |
ENSDART00000183840
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr7_+_13830052 | 1.55 |
ENSDART00000191360
|
abhd2a
|
abhydrolase domain containing 2a |
| chr1_+_12301913 | 1.54 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr19_+_26072624 | 1.49 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr21_+_15870752 | 1.46 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr3_-_40051425 | 1.44 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr7_-_26603743 | 1.43 |
ENSDART00000099003
|
plscr3b
|
phospholipid scramblase 3b |
| chr7_+_27691647 | 1.41 |
ENSDART00000079091
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr5_-_31926906 | 1.41 |
ENSDART00000187340
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr14_+_30285613 | 1.35 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr19_+_42336523 | 1.34 |
ENSDART00000151304
|
anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_23574622 | 1.30 |
ENSDART00000176012
|
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr13_+_22717939 | 1.30 |
ENSDART00000188288
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr6_+_43450221 | 1.26 |
ENSDART00000075521
|
zgc:113054
|
zgc:113054 |
| chr7_+_44802353 | 1.19 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr9_+_45227028 | 1.19 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr2_-_37353098 | 1.18 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr5_+_51227147 | 1.16 |
ENSDART00000083340
|
ubac1
|
UBA domain containing 1 |
| chr16_-_30655980 | 1.15 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr8_-_22539113 | 1.15 |
ENSDART00000183297
ENSDART00000185981 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr7_-_42206720 | 1.14 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr7_-_58244220 | 1.14 |
ENSDART00000180450
|
unm_hu7910
|
un-named hu7910 |
| chr18_+_3169579 | 1.12 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr8_-_19451487 | 1.08 |
ENSDART00000184037
|
zgc:92140
|
zgc:92140 |
| chr5_-_32505276 | 1.07 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr4_-_1776352 | 1.07 |
ENSDART00000123089
|
dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr7_+_32695954 | 1.06 |
ENSDART00000184425
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr8_-_4972916 | 1.05 |
ENSDART00000101576
|
tmem230b
|
transmembrane protein 230b |
| chr8_-_16725573 | 1.04 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr15_+_17343319 | 1.00 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr10_+_37173029 | 0.99 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr6_-_24066659 | 0.99 |
ENSDART00000164159
ENSDART00000172051 |
dr1
|
down-regulator of transcription 1 |
| chr13_-_42673978 | 0.99 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr25_-_12809361 | 0.98 |
ENSDART00000162750
|
ca5a
|
carbonic anhydrase Va |
| chr14_+_22129096 | 0.95 |
ENSDART00000132514
|
ccng1
|
cyclin G1 |
| chr5_-_65121747 | 0.95 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr15_+_43398317 | 0.94 |
ENSDART00000182528
ENSDART00000172154 ENSDART00000187688 |
actn4
|
actinin, alpha 4 |
| chr1_+_35956435 | 0.93 |
ENSDART00000085021
ENSDART00000148505 |
mmaa
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr3_-_49504023 | 0.93 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr20_+_33922339 | 0.93 |
ENSDART00000019059
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr24_+_9298198 | 0.92 |
ENSDART00000165780
|
otud1
|
OTU deubiquitinase 1 |
| chr12_+_33320504 | 0.91 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr22_-_4769140 | 0.90 |
ENSDART00000165235
|
calr3a
|
calreticulin 3a |
| chr11_+_18612166 | 0.90 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr18_+_26086803 | 0.90 |
ENSDART00000187911
|
znf710a
|
zinc finger protein 710a |
| chr2_-_37797577 | 0.90 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr6_-_4214297 | 0.89 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr13_+_22717366 | 0.89 |
ENSDART00000134122
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr7_-_48263516 | 0.88 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr12_+_48674381 | 0.88 |
ENSDART00000105309
|
uros
|
uroporphyrinogen III synthase |
| chr4_-_5018705 | 0.87 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr11_+_18612421 | 0.86 |
ENSDART00000110621
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr6_+_3373665 | 0.86 |
ENSDART00000134133
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr5_+_36666715 | 0.86 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr7_+_24881680 | 0.86 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr6_-_40922971 | 0.85 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr16_-_31351419 | 0.85 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr3_+_60828813 | 0.85 |
ENSDART00000128260
|
CABZ01087514.1
|
|
| chr2_-_32387441 | 0.84 |
ENSDART00000148202
|
ubtfl
|
upstream binding transcription factor, like |
| chr21_+_19368720 | 0.83 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr3_+_18807006 | 0.82 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr14_-_48765262 | 0.82 |
ENSDART00000166463
|
cnot6b
|
CCR4-NOT transcription complex, subunit 6b |
| chr6_+_29923593 | 0.80 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr2_-_32592084 | 0.80 |
ENSDART00000184752
|
fastk
|
Fas-activated serine/threonine kinase |
| chr6_+_6780873 | 0.79 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr22_-_14262115 | 0.79 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr6_+_44197099 | 0.79 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr12_-_35393211 | 0.78 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr9_+_22632126 | 0.78 |
ENSDART00000139434
|
etv5a
|
ets variant 5a |
| chr3_-_43650189 | 0.78 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr11_-_26701611 | 0.78 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr5_-_32505109 | 0.77 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr21_-_41070182 | 0.76 |
ENSDART00000026064
|
larsb
|
leucyl-tRNA synthetase b |
| chr16_-_42965192 | 0.76 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr16_+_32082359 | 0.76 |
ENSDART00000140794
ENSDART00000137029 |
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr25_+_20694177 | 0.74 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr6_+_40922572 | 0.74 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr8_-_49935358 | 0.73 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr13_-_25484659 | 0.73 |
ENSDART00000135321
ENSDART00000022799 |
tial1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr20_-_9123296 | 0.72 |
ENSDART00000188495
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr18_+_20034023 | 0.72 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr10_+_17747880 | 0.72 |
ENSDART00000135044
|
pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr6_-_25201810 | 0.70 |
ENSDART00000168683
|
lrrc8c
|
leucine rich repeat containing 8 VRAC subunit C |
| chr7_+_42206847 | 0.70 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr9_+_24065855 | 0.69 |
ENSDART00000161468
ENSDART00000171577 ENSDART00000172743 ENSDART00000159324 ENSDART00000079689 ENSDART00000023196 ENSDART00000101577 |
lrrfip1a
|
leucine rich repeat (in FLII) interacting protein 1a |
| chr3_+_48473346 | 0.69 |
ENSDART00000166294
|
metrnl
|
meteorin, glial cell differentiation regulator-like |
| chr18_-_12416019 | 0.69 |
ENSDART00000144799
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr16_-_31284922 | 0.69 |
ENSDART00000142638
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr9_+_22631672 | 0.69 |
ENSDART00000101770
ENSDART00000126015 ENSDART00000145005 |
etv5a
|
ets variant 5a |
| chr6_-_6248893 | 0.69 |
ENSDART00000124662
|
rtn4a
|
reticulon 4a |
| chr8_+_21146262 | 0.68 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr12_-_19119176 | 0.67 |
ENSDART00000149180
|
aco2
|
aconitase 2, mitochondrial |
| chr6_+_44197348 | 0.67 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr19_-_7321221 | 0.67 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr12_-_33770299 | 0.67 |
ENSDART00000189849
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr16_+_32082547 | 0.66 |
ENSDART00000190122
|
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr13_-_23956178 | 0.64 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr5_-_16734060 | 0.64 |
ENSDART00000035859
ENSDART00000190457 ENSDART00000179244 |
atad1a
|
ATPase family, AAA domain containing 1a |
| chr11_+_25157374 | 0.64 |
ENSDART00000019450
|
trpc4apa
|
transient receptor potential cation channel, subfamily C, member 4 associated protein a |
| chr7_+_31425976 | 0.63 |
ENSDART00000075400
|
eif3jb
|
eukaryotic translation initiation factor 3, subunit Jb |
| chr11_-_34478225 | 0.62 |
ENSDART00000189604
|
xxylt1
|
xyloside xylosyltransferase 1 |
| chr16_-_17200120 | 0.61 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr12_-_999762 | 0.61 |
ENSDART00000127003
ENSDART00000084076 ENSDART00000152425 |
mettl9
|
methyltransferase like 9 |
| chr13_-_23956361 | 0.61 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr13_-_25196758 | 0.60 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr16_-_31475904 | 0.60 |
ENSDART00000145691
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr6_+_46697710 | 0.60 |
ENSDART00000154969
|
tespa1
|
thymocyte expressed, positive selection associated 1 |
| chr9_+_29643036 | 0.60 |
ENSDART00000023210
ENSDART00000175160 |
trim13
|
tripartite motif containing 13 |
| chr9_-_28399071 | 0.59 |
ENSDART00000104317
ENSDART00000064343 |
klf7b
|
Kruppel-like factor 7b |
| chr20_+_28407720 | 0.59 |
ENSDART00000114929
|
zfyve1
|
zinc finger, FYVE domain containing 1 |
| chr13_+_6188759 | 0.59 |
ENSDART00000161062
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr1_-_53685090 | 0.58 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr23_-_27506161 | 0.57 |
ENSDART00000145007
|
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr8_+_47571211 | 0.57 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr16_+_36748538 | 0.57 |
ENSDART00000139069
|
decr1
|
2,4-dienoyl CoA reductase 1, mitochondrial |
| chr14_-_27297123 | 0.57 |
ENSDART00000173423
|
pcdh11
|
protocadherin 11 |
| chr1_+_24469313 | 0.56 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr3_-_49138004 | 0.56 |
ENSDART00000167173
|
gipc1
|
GIPC PDZ domain containing family, member 1 |
| chr5_+_37720777 | 0.55 |
ENSDART00000076083
|
cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2 |
| chr18_-_41161828 | 0.55 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr6_+_40554551 | 0.55 |
ENSDART00000017859
ENSDART00000155928 |
ddi2
|
DNA-damage inducible protein 2 |
| chr19_-_11431278 | 0.55 |
ENSDART00000109889
|
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr5_-_18911114 | 0.55 |
ENSDART00000014434
|
bri3bp
|
bri3 binding protein |
| chr20_+_41021054 | 0.54 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr20_-_40360571 | 0.54 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr5_+_61658282 | 0.53 |
ENSDART00000188878
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr21_-_27619701 | 0.51 |
ENSDART00000133441
ENSDART00000180100 |
pcnxl3
|
pecanex-like 3 (Drosophila) |
| chr7_-_30492018 | 0.51 |
ENSDART00000099639
ENSDART00000162705 ENSDART00000173663 |
adam10a
|
ADAM metallopeptidase domain 10a |
| chr5_+_51248784 | 0.51 |
ENSDART00000159571
ENSDART00000189513 ENSDART00000092285 |
msh3
|
mutS homolog 3 (E. coli) |
| chr18_+_45958375 | 0.51 |
ENSDART00000108636
|
ap2m1b
|
adaptor-related protein complex 2, mu 1 subunit, b |
| chr24_-_11076400 | 0.51 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr3_+_12593558 | 0.51 |
ENSDART00000186891
ENSDART00000159252 |
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr6_-_54348568 | 0.51 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr2_-_23349116 | 0.51 |
ENSDART00000099690
|
fam129ab
|
family with sequence similarity 129, member Ab |
| chr3_-_13546610 | 0.51 |
ENSDART00000159647
|
amdhd2
|
amidohydrolase domain containing 2 |
| chr2_+_35732652 | 0.50 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr16_+_33163858 | 0.49 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr7_-_23745984 | 0.48 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr18_-_31105391 | 0.48 |
ENSDART00000039495
|
pdcd5
|
programmed cell death 5 |
| chr7_+_7511914 | 0.47 |
ENSDART00000172848
|
clcn3
|
chloride channel 3 |
| chr20_+_43691208 | 0.46 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr21_-_26715270 | 0.46 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr25_-_8160030 | 0.45 |
ENSDART00000067159
|
tph1a
|
tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) a |
| chr11_-_11336986 | 0.45 |
ENSDART00000016677
|
zgc:77929
|
zgc:77929 |
| chr3_-_6441619 | 0.45 |
ENSDART00000157771
ENSDART00000166758 |
sumo2b
|
small ubiquitin-like modifier 2b |
| chr17_+_1496107 | 0.45 |
ENSDART00000187804
|
LO018430.1
|
|
| chr5_+_61657702 | 0.44 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_+_44346691 | 0.43 |
ENSDART00000034523
|
tars
|
threonyl-tRNA synthetase |
| chr21_-_27251112 | 0.43 |
ENSDART00000137667
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr23_-_5685023 | 0.43 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr16_+_35401543 | 0.42 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr3_+_31717291 | 0.42 |
ENSDART00000058221
|
dcaf7
|
ddb1 and cul4 associated factor 7 |
| chr3_+_45368973 | 0.39 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr7_-_49646251 | 0.39 |
ENSDART00000193674
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr13_-_31389661 | 0.38 |
ENSDART00000134630
|
zdhhc16a
|
zinc finger, DHHC-type containing 16a |
| chr15_-_506010 | 0.38 |
ENSDART00000155472
|
nudt8
|
nudix (nucleoside diphosphate linked moiety X)-type motif 8 |
| chr16_+_33987892 | 0.37 |
ENSDART00000166302
|
pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr21_+_26606741 | 0.37 |
ENSDART00000002767
|
esrra
|
estrogen-related receptor alpha |
| chr12_+_23866368 | 0.37 |
ENSDART00000188652
ENSDART00000192478 |
svila
|
supervillin a |
| chr17_-_20118145 | 0.37 |
ENSDART00000149737
ENSDART00000165606 |
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr2_-_40135942 | 0.37 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr3_+_59051503 | 0.36 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr17_+_24111392 | 0.36 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr13_+_11439486 | 0.35 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr25_+_7398689 | 0.35 |
ENSDART00000111151
|
peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr8_-_53773539 | 0.35 |
ENSDART00000098031
|
apeh
|
acylaminoacyl-peptide hydrolase |
| chr7_+_22718251 | 0.35 |
ENSDART00000027718
ENSDART00000143341 |
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr2_+_35880600 | 0.35 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr20_-_36575475 | 0.34 |
ENSDART00000062893
|
enah
|
enabled homolog (Drosophila) |
| chr9_-_52814204 | 0.34 |
ENSDART00000140771
ENSDART00000007401 |
MAP3K13
|
si:ch211-45c16.2 |
| chr25_-_27722309 | 0.34 |
ENSDART00000148121
|
zgc:153935
|
zgc:153935 |
| chr3_-_13946446 | 0.34 |
ENSDART00000171249
|
gcdhb
|
glutaryl-CoA dehydrogenase b |
| chr22_-_876506 | 0.33 |
ENSDART00000137522
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr14_+_32838110 | 0.33 |
ENSDART00000158077
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr19_-_10394931 | 0.33 |
ENSDART00000191549
|
zgc:194578
|
zgc:194578 |
| chr25_-_27722614 | 0.33 |
ENSDART00000190154
|
zgc:153935
|
zgc:153935 |
| chr23_-_35347714 | 0.33 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr1_-_23040358 | 0.33 |
ENSDART00000146871
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr8_-_23758312 | 0.33 |
ENSDART00000132659
|
INAVA
|
si:ch211-163l21.4 |
| chr1_+_13930625 | 0.32 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr8_-_17184482 | 0.32 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr16_+_7991274 | 0.31 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr21_+_12010505 | 0.30 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2ca+mef2cb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.5 | 1.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.5 | 0.5 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.5 | 1.5 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 2.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.3 | 1.5 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 2.1 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 1.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.4 | GO:0071305 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.2 | 1.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.7 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.6 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.2 | 0.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 1.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 0.7 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.2 | 0.7 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 0.7 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 0.9 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.2 | 3.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.2 | 0.5 | GO:0051096 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.2 | 1.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 1.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.6 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 2.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.9 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.7 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 1.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 1.7 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.9 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.1 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.8 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.3 | GO:0015824 | L-alanine transport(GO:0015808) proline transport(GO:0015824) |
| 0.1 | 2.1 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.5 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.6 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 1.0 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 2.2 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 0.6 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.8 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.4 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.8 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.5 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.9 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.3 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.1 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.8 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.2 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.6 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.5 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.9 | GO:0042752 | regulation of circadian rhythm(GO:0042752) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 1.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.8 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.4 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.7 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 3.5 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 1.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of BMP from receptor via BMP binding(GO:0038098) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 1.3 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0048041 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.1 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.5 | 1.5 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.4 | 1.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.4 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 1.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.0 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.5 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.4 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.1 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.5 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.0 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.4 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.7 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 2.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 2.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.6 | 1.8 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.3 | 1.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.3 | 0.8 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 1.5 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.2 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.2 | 0.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 0.7 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.2 | 0.5 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.2 | 1.0 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.2 | 0.6 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 2.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 2.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.1 | 0.9 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 1.7 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 1.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.5 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.1 | 0.6 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 4.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.3 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 2.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.5 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.5 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.1 | 0.2 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.8 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 2.0 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.9 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 2.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.7 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 1.4 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 1.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.7 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 4.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.6 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.8 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.8 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.2 | 1.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 2.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 2.0 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 0.5 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.0 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |