Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
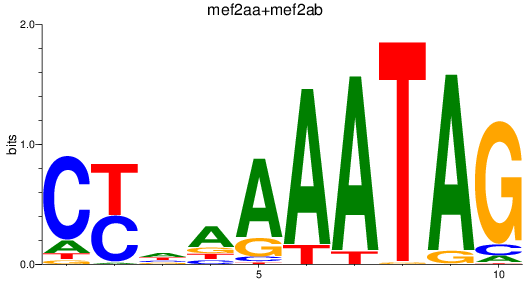
Results for mef2aa+mef2ab
Z-value: 0.33
Transcription factors associated with mef2aa+mef2ab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mef2aa
|
ENSDARG00000031756 | myocyte enhancer factor 2aa |
|
mef2ab
|
ENSDARG00000057527 | myocyte enhancer factor 2ab |
|
mef2ab
|
ENSDARG00000110771 | myocyte enhancer factor 2ab |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mef2aa | dr11_v1_chr18_+_23249519_23249519 | -0.92 | 4.1e-08 | Click! |
| mef2ab | dr11_v1_chr7_+_15659280_15659280 | 0.66 | 2.9e-03 | Click! |
Activity profile of mef2aa+mef2ab motif
Sorted Z-values of mef2aa+mef2ab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_10304981 | 0.48 |
ENSDART00000160766
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr16_-_17200120 | 0.47 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr18_-_21929426 | 0.41 |
ENSDART00000132034
ENSDART00000079050 |
nutf2
|
nuclear transport factor 2 |
| chr14_+_1240419 | 0.39 |
ENSDART00000181248
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr14_-_24332786 | 0.38 |
ENSDART00000173164
|
fam13b
|
family with sequence similarity 13, member B |
| chr14_+_1240235 | 0.36 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr10_-_8032885 | 0.35 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr9_-_24031461 | 0.34 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr17_-_50233493 | 0.32 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr21_+_12010505 | 0.31 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr20_-_40717900 | 0.30 |
ENSDART00000181663
|
cx43
|
connexin 43 |
| chr1_-_45662774 | 0.28 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr24_+_36239617 | 0.26 |
ENSDART00000062745
|
riok3
|
RIO kinase 3 (yeast) |
| chr13_-_36418921 | 0.26 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr7_-_48263516 | 0.25 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr25_+_20716176 | 0.25 |
ENSDART00000073651
|
ergic2
|
ERGIC and golgi 2 |
| chr17_-_6076266 | 0.24 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr5_-_36948586 | 0.24 |
ENSDART00000193606
|
h3f3c
|
H3 histone, family 3C |
| chr20_+_2134816 | 0.24 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr24_-_27409599 | 0.23 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr7_+_59649399 | 0.23 |
ENSDART00000123520
ENSDART00000040771 |
rpl34
|
ribosomal protein L34 |
| chr7_-_19003504 | 0.23 |
ENSDART00000157618
|
HACD4
|
si:ch73-41h24.1 |
| chr4_-_2719128 | 0.22 |
ENSDART00000184693
|
slco1c1
|
solute carrier organic anion transporter family, member 1C1 |
| chr15_+_24572926 | 0.22 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr2_+_8778490 | 0.22 |
ENSDART00000114471
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr11_-_39202915 | 0.22 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr16_+_35401543 | 0.22 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr25_+_20715950 | 0.21 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr16_+_34111919 | 0.21 |
ENSDART00000134037
ENSDART00000006061 ENSDART00000140552 |
tcea3
|
transcription elongation factor A (SII), 3 |
| chr2_+_8779164 | 0.20 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr23_-_19484160 | 0.20 |
ENSDART00000137026
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr15_-_35246742 | 0.20 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr11_+_41560792 | 0.19 |
ENSDART00000127292
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr10_-_8033468 | 0.19 |
ENSDART00000140476
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr20_+_15552657 | 0.18 |
ENSDART00000063912
|
jun
|
Jun proto-oncogene, AP-1 transcription factor subunit |
| chr4_+_22297839 | 0.18 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr14_-_33351103 | 0.18 |
ENSDART00000010019
|
upf3b
|
UPF3B, regulator of nonsense mediated mRNA decay |
| chr6_+_14301214 | 0.18 |
ENSDART00000129491
|
tmem182b
|
transmembrane protein 182b |
| chr15_+_5923851 | 0.18 |
ENSDART00000152520
ENSDART00000145827 ENSDART00000121529 |
sh3bgr
|
SH3 domain binding glutamate-rich protein |
| chr10_-_26512742 | 0.18 |
ENSDART00000135951
|
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr15_-_34668485 | 0.17 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr10_-_17170086 | 0.17 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr1_+_7546259 | 0.17 |
ENSDART00000015732
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr3_+_60761811 | 0.16 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr25_+_11389220 | 0.16 |
ENSDART00000187787
|
AGBL1
|
si:ch73-141f14.1 |
| chr7_+_40094081 | 0.16 |
ENSDART00000186054
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr6_+_12462079 | 0.16 |
ENSDART00000192029
ENSDART00000065385 |
nr4a2b
|
nuclear receptor subfamily 4, group A, member 2b |
| chr25_+_32530976 | 0.15 |
ENSDART00000156190
ENSDART00000103324 |
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr18_+_40354998 | 0.15 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr10_-_26512993 | 0.15 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr20_-_26066020 | 0.15 |
ENSDART00000078559
|
myct1a
|
myc target 1a |
| chr2_-_8611675 | 0.15 |
ENSDART00000138223
|
si:ch211-71m22.1
|
si:ch211-71m22.1 |
| chr23_-_9855627 | 0.15 |
ENSDART00000180159
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr7_+_7048245 | 0.15 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr11_+_3501669 | 0.15 |
ENSDART00000160808
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr9_-_24209083 | 0.15 |
ENSDART00000134599
|
zgc:153521
|
zgc:153521 |
| chr1_-_22652424 | 0.15 |
ENSDART00000036797
|
uchl1
|
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr25_+_14087045 | 0.15 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr7_+_40093662 | 0.15 |
ENSDART00000053011
|
si:ch73-174h16.4
|
si:ch73-174h16.4 |
| chr12_+_6041575 | 0.14 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr20_+_16881883 | 0.14 |
ENSDART00000130107
|
nfkbiaa
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha a |
| chr21_-_25272317 | 0.14 |
ENSDART00000170556
ENSDART00000163277 |
nfrkb
|
nuclear factor related to kappaB binding protein |
| chr5_-_41709234 | 0.14 |
ENSDART00000083656
|
atxn2
|
ataxin 2 |
| chr12_+_27537357 | 0.14 |
ENSDART00000136212
|
etv4
|
ets variant 4 |
| chr3_+_32620575 | 0.14 |
ENSDART00000151340
|
mmp25a
|
matrix metallopeptidase 25a |
| chr1_-_8917902 | 0.14 |
ENSDART00000137900
|
grin2ab
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A, b |
| chr7_-_30143092 | 0.13 |
ENSDART00000173636
|
frmd5
|
FERM domain containing 5 |
| chr21_-_5799122 | 0.13 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr14_-_36799280 | 0.13 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr7_+_6969909 | 0.13 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr20_-_7000225 | 0.13 |
ENSDART00000100098
|
adcy1a
|
adenylate cyclase 1a |
| chr20_-_47732703 | 0.12 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr16_-_36798783 | 0.12 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr23_+_12134839 | 0.12 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr19_-_7450796 | 0.12 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr14_+_23520986 | 0.12 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr11_+_44503774 | 0.12 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr6_-_10752937 | 0.12 |
ENSDART00000135093
|
ola1
|
Obg-like ATPase 1 |
| chr16_-_28878080 | 0.12 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr12_+_26632448 | 0.11 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_+_21331688 | 0.11 |
ENSDART00000128014
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr14_+_30910114 | 0.11 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr17_-_50234004 | 0.11 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr9_-_23891102 | 0.11 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr11_-_7139555 | 0.10 |
ENSDART00000016472
|
bmp7a
|
bone morphogenetic protein 7a |
| chr20_-_2134620 | 0.10 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr14_+_23518110 | 0.10 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr17_+_28340138 | 0.10 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr8_+_22327251 | 0.10 |
ENSDART00000075129
|
lrrc47
|
leucine rich repeat containing 47 |
| chr3_-_52674089 | 0.10 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr3_-_53114299 | 0.10 |
ENSDART00000109390
|
AL954361.1
|
|
| chr25_+_17862338 | 0.10 |
ENSDART00000151853
ENSDART00000090867 |
btbd10a
|
BTB (POZ) domain containing 10a |
| chr14_-_22113600 | 0.10 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr5_+_36932718 | 0.10 |
ENSDART00000037879
|
crx
|
cone-rod homeobox |
| chr5_+_45677781 | 0.10 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr16_+_25245857 | 0.09 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr6_-_13891727 | 0.09 |
ENSDART00000065356
|
desmb
|
desmin b |
| chr2_-_3678029 | 0.09 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr12_+_28910762 | 0.09 |
ENSDART00000076342
ENSDART00000160939 ENSDART00000076572 |
rnf40
|
ring finger protein 40 |
| chr12_-_6905375 | 0.09 |
ENSDART00000152322
|
pcdh15b
|
protocadherin-related 15b |
| chr17_+_44441042 | 0.09 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr3_+_28953274 | 0.09 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr19_-_3874986 | 0.09 |
ENSDART00000161830
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr19_+_5291935 | 0.09 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr3_-_61205711 | 0.09 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr2_+_27403300 | 0.09 |
ENSDART00000099180
|
elovl8a
|
ELOVL fatty acid elongase 8a |
| chr14_-_24111292 | 0.09 |
ENSDART00000186611
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr14_-_33177935 | 0.09 |
ENSDART00000180583
ENSDART00000078856 |
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr8_+_52637507 | 0.08 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr19_+_30990815 | 0.08 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr1_-_11075403 | 0.08 |
ENSDART00000102903
ENSDART00000170290 |
dmd
|
dystrophin |
| chr8_-_42594380 | 0.08 |
ENSDART00000140126
ENSDART00000135238 ENSDART00000192764 |
dok2
|
docking protein 2 |
| chr16_-_43356018 | 0.08 |
ENSDART00000181683
|
FO704821.1
|
|
| chr12_+_6002715 | 0.08 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr25_-_29988352 | 0.08 |
ENSDART00000067059
|
fam19a5b
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5b |
| chr4_+_14343706 | 0.08 |
ENSDART00000142845
|
prl2
|
prolactin 2 |
| chr7_-_49646251 | 0.08 |
ENSDART00000193674
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr19_+_2590182 | 0.08 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr10_+_17714866 | 0.08 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr25_+_25508495 | 0.07 |
ENSDART00000150719
|
phrf1
|
PHD and ring finger domains 1 |
| chr10_-_17232372 | 0.07 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr1_-_37383741 | 0.07 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr4_+_5848229 | 0.07 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr17_-_15600455 | 0.07 |
ENSDART00000110272
ENSDART00000156911 |
si:ch211-266g18.9
|
si:ch211-266g18.9 |
| chr13_+_22480857 | 0.07 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr4_+_76353418 | 0.07 |
ENSDART00000171289
|
si:ch73-158p21.2
|
si:ch73-158p21.2 |
| chr5_-_38170996 | 0.07 |
ENSDART00000145805
|
si:ch211-284e13.12
|
si:ch211-284e13.12 |
| chr3_-_42981739 | 0.07 |
ENSDART00000167844
|
mafk
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
| chr6_-_49873020 | 0.07 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr20_+_46371458 | 0.07 |
ENSDART00000152912
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr12_-_15584479 | 0.07 |
ENSDART00000150134
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr2_-_10098191 | 0.07 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr2_-_15324837 | 0.07 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr5_-_72125551 | 0.07 |
ENSDART00000149412
|
smyd1a
|
SET and MYND domain containing 1a |
| chr22_+_38301365 | 0.07 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr5_-_9948497 | 0.07 |
ENSDART00000183196
|
tor4ab
|
torsin family 4, member Ab |
| chr4_-_16853464 | 0.06 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr16_-_17541890 | 0.06 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr12_-_4781801 | 0.06 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr7_+_26716321 | 0.06 |
ENSDART00000189750
|
cd82a
|
CD82 molecule a |
| chr23_+_14097508 | 0.06 |
ENSDART00000087280
|
cacnb3a
|
calcium channel, voltage-dependent, beta 3a |
| chr23_+_6077503 | 0.06 |
ENSDART00000081714
ENSDART00000139834 |
mybpha
|
myosin binding protein Ha |
| chr16_+_29663809 | 0.06 |
ENSDART00000191336
|
tmod4
|
tropomodulin 4 (muscle) |
| chr17_-_14671098 | 0.06 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr1_+_10378706 | 0.06 |
ENSDART00000046283
ENSDART00000103535 ENSDART00000132076 |
dachb
|
dachshund b |
| chr12_+_18718065 | 0.06 |
ENSDART00000152935
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr9_-_23147026 | 0.06 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr6_+_27896657 | 0.06 |
ENSDART00000079419
|
cep63
|
centrosomal protein 63 |
| chr12_-_30338779 | 0.06 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr21_+_40589770 | 0.06 |
ENSDART00000164650
ENSDART00000161584 ENSDART00000161108 |
pdk3b
|
pyruvate dehydrogenase kinase, isozyme 3b |
| chr16_+_5774977 | 0.06 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr6_-_18698456 | 0.06 |
ENSDART00000166587
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr15_-_28802140 | 0.06 |
ENSDART00000156120
ENSDART00000156708 ENSDART00000122536 ENSDART00000155008 ENSDART00000153825 |
cd3eap
|
CD3e molecule, epsilon associated protein |
| chr22_-_20761759 | 0.06 |
ENSDART00000013803
|
amh
|
anti-Mullerian hormone |
| chr8_+_18010978 | 0.06 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr1_-_37383539 | 0.06 |
ENSDART00000127579
|
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr4_+_13449775 | 0.06 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr1_-_9195629 | 0.06 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr15_+_37412883 | 0.06 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr6_+_27897313 | 0.06 |
ENSDART00000128935
|
cep63
|
centrosomal protein 63 |
| chr17_+_48314724 | 0.06 |
ENSDART00000125617
|
smoc1
|
SPARC related modular calcium binding 1 |
| chr11_-_1948784 | 0.06 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr21_-_40083432 | 0.06 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr12_+_27141140 | 0.06 |
ENSDART00000136415
|
hoxb1b
|
homeobox B1b |
| chr7_+_20563305 | 0.05 |
ENSDART00000169661
|
si:dkey-19b23.10
|
si:dkey-19b23.10 |
| chr16_-_32837806 | 0.05 |
ENSDART00000003997
|
MCHR2
|
si:dkey-165n16.5 |
| chr19_+_43017931 | 0.05 |
ENSDART00000132213
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr17_-_4395373 | 0.05 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr5_+_21922534 | 0.05 |
ENSDART00000148092
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr2_-_28102264 | 0.05 |
ENSDART00000013638
|
cdh6
|
cadherin 6 |
| chr25_-_15040369 | 0.05 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr1_+_17376922 | 0.05 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr23_+_33413804 | 0.05 |
ENSDART00000142328
|
rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr21_-_131236 | 0.05 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr21_+_20383837 | 0.05 |
ENSDART00000026430
|
hspb11
|
heat shock protein, alpha-crystallin-related, b11 |
| chr12_-_26064105 | 0.05 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr20_+_6630540 | 0.05 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr21_-_13972745 | 0.05 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr24_+_19986432 | 0.05 |
ENSDART00000184402
|
CR381686.5
|
|
| chr24_+_34089977 | 0.05 |
ENSDART00000157466
|
asb10
|
ankyrin repeat and SOCS box containing 10 |
| chr3_+_52442102 | 0.05 |
ENSDART00000155455
|
si:ch211-191c10.1
|
si:ch211-191c10.1 |
| chr15_+_15856178 | 0.05 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr4_+_6032640 | 0.05 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr7_+_50464500 | 0.05 |
ENSDART00000191356
|
serinc4
|
serine incorporator 4 |
| chr23_+_17981127 | 0.05 |
ENSDART00000012571
ENSDART00000145200 |
chia.6
|
chitinase, acidic.6 |
| chr3_-_50046004 | 0.05 |
ENSDART00000109544
|
si:ch1073-100f3.2
|
si:ch1073-100f3.2 |
| chr2_-_5356686 | 0.05 |
ENSDART00000124290
|
MFN1
|
mitofusin 1 |
| chr8_-_27687095 | 0.05 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr11_+_25504215 | 0.05 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr13_-_50565338 | 0.05 |
ENSDART00000062684
|
blnk
|
B cell linker |
| chr4_-_72036820 | 0.05 |
ENSDART00000168454
|
pyroxd1
|
pyridine nucleotide-disulphide oxidoreductase domain 1 |
| chr8_-_32497815 | 0.05 |
ENSDART00000122359
|
si:dkey-164f24.2
|
si:dkey-164f24.2 |
| chr25_-_7764083 | 0.05 |
ENSDART00000179800
ENSDART00000181858 |
phf21ab
|
PHD finger protein 21Ab |
| chr21_+_26606741 | 0.05 |
ENSDART00000002767
|
esrra
|
estrogen-related receptor alpha |
| chr3_-_56889052 | 0.05 |
ENSDART00000188412
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr6_-_18199062 | 0.05 |
ENSDART00000167513
|
ppp1r27b
|
protein phosphatase 1, regulatory subunit 27b |
| chr19_+_43017628 | 0.05 |
ENSDART00000182608
ENSDART00000015632 |
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr23_+_33907899 | 0.04 |
ENSDART00000159445
|
cs
|
citrate synthase |
| chr3_-_17871846 | 0.04 |
ENSDART00000074478
ENSDART00000187941 |
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr14_-_2318590 | 0.04 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mef2aa+mef2ab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.3 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.0 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.0 | 0.5 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.1 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.1 | GO:0090200 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.2 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.0 | GO:0042420 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0003418 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.0 | 0.3 | GO:0007634 | optokinetic behavior(GO:0007634) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0050780 | mu-type opioid receptor binding(GO:0031852) dopamine receptor binding(GO:0050780) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.2 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |