Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
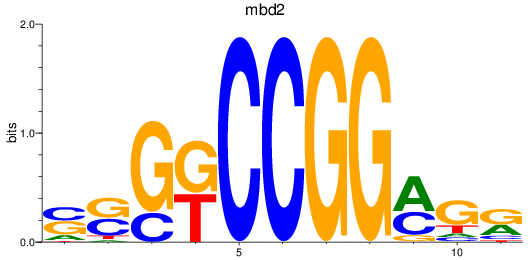
Results for mbd2
Z-value: 0.43
Transcription factors associated with mbd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mbd2
|
ENSDARG00000075952 | methyl-CpG binding domain protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mbd2 | dr11_v1_chr5_-_1047222_1047222 | -0.35 | 1.6e-01 | Click! |
Activity profile of mbd2 motif
Sorted Z-values of mbd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_127558 | 1.06 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr19_-_42045372 | 0.88 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr10_+_16225870 | 0.82 |
ENSDART00000164647
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr18_-_127873 | 0.69 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr19_+_14352332 | 0.59 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr10_+_16036573 | 0.59 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr22_-_36829006 | 0.58 |
ENSDART00000170256
ENSDART00000086064 |
map1sa
|
microtubule-associated protein 1Sa |
| chr2_-_56635744 | 0.57 |
ENSDART00000167790
ENSDART00000168160 |
pip5k1cb
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma b |
| chr10_+_16036246 | 0.57 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr2_-_32738535 | 0.55 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr3_+_16664212 | 0.53 |
ENSDART00000013816
|
zgc:55558
|
zgc:55558 |
| chr7_-_9556354 | 0.53 |
ENSDART00000127974
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr14_+_44860335 | 0.53 |
ENSDART00000091620
ENSDART00000173043 ENSDART00000091625 |
atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr15_-_41677689 | 0.52 |
ENSDART00000187063
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr8_-_20230802 | 0.51 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr8_-_20230559 | 0.50 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr18_+_3579829 | 0.50 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr23_-_9807546 | 0.50 |
ENSDART00000136740
ENSDART00000004474 |
mapre1b
|
microtubule-associated protein, RP/EB family, member 1b |
| chr22_-_506522 | 0.47 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr8_+_8643901 | 0.46 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr24_-_35699595 | 0.44 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_+_6962271 | 0.44 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr4_-_25796848 | 0.43 |
ENSDART00000122881
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr22_-_3564563 | 0.42 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr8_+_23861461 | 0.40 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr24_-_35699444 | 0.38 |
ENSDART00000166567
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_+_2452672 | 0.36 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr5_+_393738 | 0.35 |
ENSDART00000161456
|
june
|
JunE proto-oncogene, AP-1 transcription factor subunit |
| chr12_-_3773869 | 0.35 |
ENSDART00000092983
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr14_-_30983011 | 0.34 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr15_+_11693624 | 0.34 |
ENSDART00000193630
ENSDART00000161930 |
strn4
|
striatin, calmodulin binding protein 4 |
| chr20_+_46699021 | 0.33 |
ENSDART00000167398
ENSDART00000029894 |
wdr26b
|
WD repeat domain 26b |
| chr22_+_10606863 | 0.32 |
ENSDART00000147975
|
rad54l2
|
RAD54 like 2 |
| chr1_+_52792439 | 0.32 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr10_+_16225553 | 0.31 |
ENSDART00000129844
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr22_-_5099824 | 0.31 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr20_+_34502606 | 0.30 |
ENSDART00000139739
|
gorab
|
golgin, rab6-interacting |
| chr3_-_5228137 | 0.30 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr22_+_10606573 | 0.30 |
ENSDART00000192638
|
rad54l2
|
RAD54 like 2 |
| chr18_-_44847855 | 0.29 |
ENSDART00000086823
|
srpr
|
signal recognition particle receptor (docking protein) |
| chr18_+_18879733 | 0.28 |
ENSDART00000019581
|
arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila) |
| chr12_+_5708400 | 0.28 |
ENSDART00000017191
|
dlx3b
|
distal-less homeobox 3b |
| chr20_+_40457599 | 0.27 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr4_+_279669 | 0.27 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr1_-_69444 | 0.27 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr6_-_21726758 | 0.26 |
ENSDART00000083085
|
mtmr14
|
myotubularin related protein 14 |
| chr20_-_44055095 | 0.25 |
ENSDART00000125898
ENSDART00000082265 |
runx2b
|
runt-related transcription factor 2b |
| chr13_-_51922290 | 0.24 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr23_-_17450746 | 0.23 |
ENSDART00000145399
ENSDART00000136457 ENSDART00000133125 ENSDART00000145719 ENSDART00000147524 ENSDART00000005366 ENSDART00000104680 |
tpd52l2b
|
tumor protein D52-like 2b |
| chr16_+_38940758 | 0.23 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr20_+_18943406 | 0.23 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr25_+_20272145 | 0.23 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr2_+_50062165 | 0.22 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr17_-_10738001 | 0.21 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr22_-_12862415 | 0.21 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr10_+_6013076 | 0.21 |
ENSDART00000167613
ENSDART00000159216 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr2_+_56139941 | 0.20 |
ENSDART00000164741
|
pgpep1
|
pyroglutamyl-peptidase I |
| chr21_-_45685063 | 0.20 |
ENSDART00000162422
|
sar1b
|
secretion associated, Ras related GTPase 1B |
| chr13_+_51710725 | 0.20 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr10_-_28193642 | 0.18 |
ENSDART00000019050
|
rps6kb1a
|
ribosomal protein S6 kinase b, polypeptide 1a |
| chr16_+_27564270 | 0.18 |
ENSDART00000140460
|
tmem67
|
transmembrane protein 67 |
| chr13_-_45155284 | 0.18 |
ENSDART00000161144
|
runx3
|
runt-related transcription factor 3 |
| chr23_-_7052362 | 0.17 |
ENSDART00000127702
ENSDART00000192468 |
prpf6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) |
| chr9_+_711822 | 0.17 |
ENSDART00000136627
|
ybey
|
ybeY metallopeptidase |
| chr13_-_27620815 | 0.17 |
ENSDART00000139904
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr9_+_711638 | 0.16 |
ENSDART00000191964
|
ybey
|
ybeY metallopeptidase |
| chr12_-_287633 | 0.16 |
ENSDART00000113335
|
map2k4b
|
mitogen-activated protein kinase kinase 4b |
| chr7_-_7797654 | 0.15 |
ENSDART00000084503
ENSDART00000192779 ENSDART00000173079 |
trmt10b
|
tRNA methyltransferase 10B |
| chr5_-_23485161 | 0.15 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr17_-_8673278 | 0.14 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr8_-_41519234 | 0.14 |
ENSDART00000167283
ENSDART00000180666 |
golga1
|
golgin A1 |
| chr13_+_4888351 | 0.13 |
ENSDART00000145940
|
micu1
|
mitochondrial calcium uptake 1 |
| chr17_+_10738126 | 0.13 |
ENSDART00000051527
|
tbpl2
|
TATA box binding protein like 2 |
| chr19_-_45534392 | 0.13 |
ENSDART00000163920
|
trps1
|
trichorhinophalangeal syndrome I |
| chr6_-_31336317 | 0.11 |
ENSDART00000193927
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr13_-_44782462 | 0.11 |
ENSDART00000141298
ENSDART00000099990 |
btbd9
|
BTB (POZ) domain containing 9 |
| chr14_-_45967712 | 0.11 |
ENSDART00000043751
ENSDART00000141357 |
macrod1
|
MACRO domain containing 1 |
| chr9_-_21936841 | 0.09 |
ENSDART00000144843
|
lmo7a
|
LIM domain 7a |
| chr17_-_26604549 | 0.09 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr16_-_27161410 | 0.09 |
ENSDART00000177503
|
LO017771.1
|
|
| chr15_-_3078600 | 0.08 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr25_+_34407740 | 0.07 |
ENSDART00000012677
|
OTUD7A
|
si:dkey-37f18.2 |
| chr15_+_30158652 | 0.07 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr22_-_2929127 | 0.07 |
ENSDART00000123730
ENSDART00000046678 |
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr19_-_18313303 | 0.07 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr25_+_34407529 | 0.07 |
ENSDART00000156751
|
OTUD7A
|
si:dkey-37f18.2 |
| chr22_+_1853999 | 0.07 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr7_-_35314347 | 0.07 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr4_+_68562464 | 0.06 |
ENSDART00000192954
|
BX548011.4
|
|
| chr17_+_6382275 | 0.06 |
ENSDART00000056324
|
CR628323.2
|
|
| chr1_-_22803147 | 0.06 |
ENSDART00000086867
|
tapt1b
|
transmembrane anterior posterior transformation 1b |
| chr23_+_39331216 | 0.05 |
ENSDART00000160957
|
kcng1
|
potassium voltage-gated channel, subfamily G, member 1 |
| chr4_-_77557279 | 0.04 |
ENSDART00000180113
|
AL935186.10
|
|
| chr7_+_53755054 | 0.04 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr7_+_53754653 | 0.04 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr14_+_94603 | 0.04 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr19_+_56351 | 0.04 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr21_-_12272543 | 0.03 |
ENSDART00000081510
ENSDART00000151297 |
celf4
|
CUGBP, Elav-like family member 4 |
| chr2_+_35854242 | 0.03 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr9_+_24159725 | 0.03 |
ENSDART00000137756
|
hecw2a
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2a |
| chr6_-_1874664 | 0.02 |
ENSDART00000007972
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr13_-_133520 | 0.02 |
ENSDART00000180874
|
FO904997.1
|
|
| chr6_+_49771626 | 0.02 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr14_+_743346 | 0.02 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr18_-_46763170 | 0.02 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr19_+_770300 | 0.02 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr6_-_19664848 | 0.02 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr17_-_50010121 | 0.02 |
ENSDART00000122747
|
tmem30aa
|
transmembrane protein 30Aa |
| chr6_-_19665114 | 0.01 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr5_-_8817458 | 0.01 |
ENSDART00000191098
|
fgf10b
|
fibroblast growth factor 10b |
| chr11_-_27738489 | 0.01 |
ENSDART00000181612
|
fam120a
|
family with sequence similarity 120A |
| chr22_+_35275206 | 0.01 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr3_-_17716322 | 0.01 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr17_-_52636937 | 0.01 |
ENSDART00000084268
|
fam98b
|
family with sequence similarity 98, member B |
| chr13_-_40120252 | 0.01 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr20_+_812012 | 0.00 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of mbd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.3 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.4 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.1 | 1.1 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.2 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.2 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.3 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.6 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.2 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 1.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.2 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |