Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
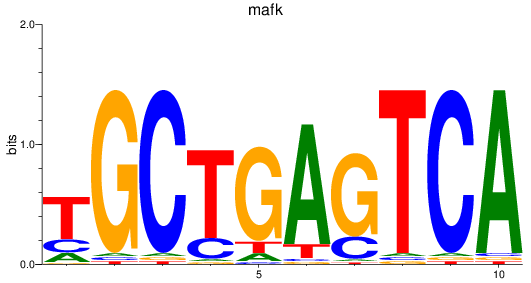
Results for mafk
Z-value: 1.28
Transcription factors associated with mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafk
|
ENSDARG00000100947 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafk | dr11_v1_chr3_-_42981739_42981739 | -0.53 | 2.5e-02 | Click! |
Activity profile of mafk motif
Sorted Z-values of mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_30960470 | 4.61 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr18_+_7611298 | 4.04 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_66355153 | 3.94 |
ENSDART00000082745
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr18_+_7456597 | 3.90 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr24_+_24923166 | 3.76 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr13_+_2908764 | 3.13 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr3_-_26017592 | 3.00 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr11_+_26609110 | 2.90 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr6_+_48348415 | 2.85 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr12_+_1455147 | 2.80 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr1_-_52498146 | 2.79 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr13_-_37642890 | 2.77 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr2_+_10709557 | 2.74 |
ENSDART00000183118
ENSDART00000109723 |
evi5a
|
ecotropic viral integration site 5a |
| chr11_-_24681292 | 2.65 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr23_+_9560991 | 2.59 |
ENSDART00000081433
ENSDART00000131594 ENSDART00000130069 ENSDART00000138601 |
adrm1
|
adhesion regulating molecule 1 |
| chr3_-_26017831 | 2.50 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr19_-_6873107 | 2.49 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr5_+_62723233 | 2.40 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr22_+_1049367 | 2.39 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr17_+_7524389 | 2.32 |
ENSDART00000067579
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr10_+_26747755 | 2.29 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr8_-_13999293 | 2.27 |
ENSDART00000142098
|
si:dkeyp-110g5.4
|
si:dkeyp-110g5.4 |
| chr1_+_44260085 | 2.25 |
ENSDART00000171416
|
si:dkey-59p5.1
|
si:dkey-59p5.1 |
| chr3_+_7617353 | 2.20 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr3_-_18030938 | 2.17 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr15_+_37559570 | 1.98 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr1_+_44360973 | 1.97 |
ENSDART00000167568
|
si:ch211-165a10.5
|
si:ch211-165a10.5 |
| chr23_+_3731375 | 1.94 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr8_-_50259448 | 1.94 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr18_-_17020231 | 1.86 |
ENSDART00000129146
|
tbc1d15
|
TBC1 domain family, member 15 |
| chr21_-_13225402 | 1.85 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr3_-_58831683 | 1.84 |
ENSDART00000110292
|
IGKV1D-12
|
immunoglobulin kappa variable 1D-12 |
| chr10_-_322769 | 1.83 |
ENSDART00000165244
|
akt2l
|
v-akt murine thymoma viral oncogene homolog 2, like |
| chr1_+_57311901 | 1.82 |
ENSDART00000149397
|
mycbpap
|
mycbp associated protein |
| chr1_-_52497834 | 1.81 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr1_+_44249616 | 1.81 |
ENSDART00000164173
|
si:ch211-165a10.10
|
si:ch211-165a10.10 |
| chr5_-_19052184 | 1.81 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr6_+_12853655 | 1.80 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr23_+_19813677 | 1.80 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr4_-_390431 | 1.80 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr24_+_38522254 | 1.78 |
ENSDART00000156189
|
si:ch1073-66l23.1
|
si:ch1073-66l23.1 |
| chr21_-_25756119 | 1.77 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr1_+_9994811 | 1.76 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr21_-_5393125 | 1.76 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr12_+_10163585 | 1.75 |
ENSDART00000106191
|
psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr7_+_22657566 | 1.71 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr6_+_33885828 | 1.71 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr22_-_36934040 | 1.69 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr19_-_10881141 | 1.69 |
ENSDART00000162793
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr19_-_32600823 | 1.67 |
ENSDART00000134149
ENSDART00000187858 |
zgc:91944
|
zgc:91944 |
| chr7_+_21331688 | 1.66 |
ENSDART00000128014
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr3_-_54992934 | 1.63 |
ENSDART00000053095
ENSDART00000145766 |
rhbdf1a
|
rhomboid 5 homolog 1a (Drosophila) |
| chr19_-_6385594 | 1.63 |
ENSDART00000104950
|
atp1a3a
|
ATPase Na+/K+ transporting subunit alpha 3a |
| chr19_-_41404870 | 1.62 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr12_-_19279103 | 1.60 |
ENSDART00000186669
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr20_-_1314537 | 1.57 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr14_-_26397849 | 1.57 |
ENSDART00000148140
|
pimr212
|
Pim proto-oncogene, serine/threonine kinase, related 212 |
| chr12_-_1951233 | 1.56 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr16_+_11029762 | 1.56 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr6_-_31364475 | 1.55 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr7_+_756942 | 1.51 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr5_+_37087583 | 1.50 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr19_-_10881486 | 1.50 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr3_+_48234445 | 1.48 |
ENSDART00000161419
|
tbcd
|
tubulin folding cofactor D |
| chr22_-_557965 | 1.46 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr16_-_13613475 | 1.46 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr1_+_6640437 | 1.44 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr6_-_138392 | 1.41 |
ENSDART00000148974
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr6_-_35446110 | 1.38 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr11_+_45255774 | 1.38 |
ENSDART00000172838
|
aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr7_-_38644560 | 1.38 |
ENSDART00000114934
|
c6ast3
|
six-cysteine containing astacin protease 3 |
| chr3_-_8765165 | 1.38 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr5_-_27867657 | 1.37 |
ENSDART00000112495
|
tcima
|
transcriptional and immune response regulator a |
| chr20_+_34326874 | 1.35 |
ENSDART00000061659
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr21_+_3960583 | 1.34 |
ENSDART00000149788
|
setx
|
senataxin |
| chr11_-_40128722 | 1.34 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr3_+_26064091 | 1.33 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr6_+_15762647 | 1.30 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr19_+_8506178 | 1.29 |
ENSDART00000189689
|
s100a10a
|
S100 calcium binding protein A10a |
| chr4_-_2545310 | 1.29 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr14_+_11457500 | 1.27 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr19_+_19786117 | 1.27 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr25_-_15214161 | 1.26 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr20_-_1314355 | 1.25 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr13_+_35746440 | 1.23 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr10_+_28428222 | 1.23 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr6_-_52723901 | 1.22 |
ENSDART00000033949
|
oser1
|
oxidative stress responsive serine-rich 1 |
| chr6_-_138603 | 1.21 |
ENSDART00000148911
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr19_+_7115223 | 1.20 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr22_+_31059919 | 1.20 |
ENSDART00000077063
|
sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr1_-_20928772 | 1.18 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr12_+_48216662 | 1.17 |
ENSDART00000187369
|
lrrc20
|
leucine rich repeat containing 20 |
| chr11_+_13630107 | 1.15 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr5_+_4298636 | 1.14 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr1_-_8652648 | 1.12 |
ENSDART00000138324
ENSDART00000141407 ENSDART00000054987 |
actb1
|
actin, beta 1 |
| chr5_+_65946222 | 1.11 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr7_-_50917255 | 1.10 |
ENSDART00000022918
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr24_-_37680917 | 1.10 |
ENSDART00000131342
|
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr13_-_23007813 | 1.10 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr16_-_21785261 | 1.08 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr22_-_13857729 | 1.08 |
ENSDART00000177971
|
s100b
|
S100 calcium binding protein, beta (neural) |
| chr8_+_41038141 | 1.08 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr1_+_44239558 | 1.08 |
ENSDART00000100335
|
si:dkey-59p5.2
|
si:dkey-59p5.2 |
| chr7_+_24114694 | 1.07 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr19_-_7115229 | 1.06 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr5_+_64907368 | 1.05 |
ENSDART00000122863
|
ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr12_-_31457301 | 1.03 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr11_-_10770053 | 1.02 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr7_+_20475788 | 1.02 |
ENSDART00000171155
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr8_-_23573084 | 1.00 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr11_-_3334248 | 0.99 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr14_+_46216703 | 0.97 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr12_-_22540943 | 0.96 |
ENSDART00000172310
|
zbtb4
|
zinc finger and BTB domain containing 4 |
| chr20_-_25626198 | 0.94 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr25_+_10485103 | 0.93 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr25_-_35045250 | 0.92 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr15_-_39820491 | 0.92 |
ENSDART00000097134
|
robo1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr12_-_38548299 | 0.92 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr1_-_1894722 | 0.91 |
ENSDART00000165669
|
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr24_-_42148339 | 0.91 |
ENSDART00000112680
|
rmdn1
|
regulator of microtubule dynamics 1 |
| chr12_+_2446837 | 0.90 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr2_-_24962002 | 0.90 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr25_+_22643954 | 0.89 |
ENSDART00000182843
ENSDART00000121791 |
ush1c
|
Usher syndrome 1C |
| chr21_-_20832482 | 0.89 |
ENSDART00000191928
|
c6
|
complement component 6 |
| chr16_+_24721914 | 0.89 |
ENSDART00000109459
|
smg9
|
smg9 nonsense mediated mRNA decay factor |
| chr12_-_28363111 | 0.88 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr8_-_17516448 | 0.88 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr22_+_37631234 | 0.87 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr20_-_31905968 | 0.87 |
ENSDART00000142806
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr7_-_25697285 | 0.87 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr11_+_42730639 | 0.87 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr16_-_16182319 | 0.87 |
ENSDART00000103815
|
stmn2a
|
stathmin 2a |
| chr24_-_16917086 | 0.87 |
ENSDART00000110715
|
cmbl
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr10_+_466926 | 0.87 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr19_+_31771270 | 0.86 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr19_-_10243148 | 0.85 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr22_+_38192568 | 0.84 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr2_-_3038904 | 0.84 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr7_+_63325819 | 0.84 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr22_+_2861734 | 0.83 |
ENSDART00000140578
|
si:dkey-20i20.2
|
si:dkey-20i20.2 |
| chr6_-_39653972 | 0.83 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr20_-_19422496 | 0.82 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr23_+_28128453 | 0.82 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr1_+_144284 | 0.81 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr18_+_2189211 | 0.80 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr16_-_48673938 | 0.80 |
ENSDART00000156969
|
notchl
|
notch homolog, like |
| chr22_+_37631034 | 0.80 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr6_+_41463786 | 0.79 |
ENSDART00000065006
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr7_+_33172066 | 0.79 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr2_-_16562505 | 0.79 |
ENSDART00000156406
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr10_-_300000 | 0.78 |
ENSDART00000183273
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr23_+_17865953 | 0.78 |
ENSDART00000014723
ENSDART00000140302 ENSDART00000144800 |
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr19_+_26828502 | 0.77 |
ENSDART00000183654
ENSDART00000126006 ENSDART00000185341 |
FP085398.1
|
|
| chr24_+_38534550 | 0.76 |
ENSDART00000105677
|
zgc:154125
|
zgc:154125 |
| chr16_-_30880236 | 0.75 |
ENSDART00000035583
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr9_+_28688574 | 0.75 |
ENSDART00000101319
|
zgc:162396
|
zgc:162396 |
| chr4_-_9728730 | 0.75 |
ENSDART00000150265
|
mical3b
|
microtubule associated monooxygenase, calponin and LIM domain containing 3b |
| chr18_-_43866001 | 0.75 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr9_-_2573121 | 0.75 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr18_+_14477740 | 0.74 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr22_+_19528851 | 0.74 |
ENSDART00000145079
|
si:dkey-78l4.13
|
si:dkey-78l4.13 |
| chr22_-_31059670 | 0.74 |
ENSDART00000022445
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr4_-_16334362 | 0.74 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr24_+_16905188 | 0.73 |
ENSDART00000066760
|
cct5
|
chaperonin containing TCP1, subunit 5 (epsilon) |
| chr3_-_30158395 | 0.72 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr21_-_23110841 | 0.72 |
ENSDART00000147896
ENSDART00000003076 ENSDART00000184925 ENSDART00000190386 |
usp28
|
ubiquitin specific peptidase 28 |
| chr18_-_5692292 | 0.72 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr6_-_8498908 | 0.72 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr7_+_62004048 | 0.72 |
ENSDART00000181818
|
smim20
|
small integral membrane protein 20 |
| chr14_-_17588345 | 0.72 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr23_-_19484160 | 0.71 |
ENSDART00000137026
|
fam208ab
|
family with sequence similarity 208, member Ab |
| chr19_+_31404686 | 0.70 |
ENSDART00000078459
|
pip4p2
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 2 |
| chr20_-_39271844 | 0.70 |
ENSDART00000192708
|
clu
|
clusterin |
| chr14_+_49395437 | 0.69 |
ENSDART00000172850
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr6_-_426041 | 0.69 |
ENSDART00000162789
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr9_+_34232503 | 0.69 |
ENSDART00000132836
|
nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr6_-_43882696 | 0.69 |
ENSDART00000064938
|
foxp1b
|
forkhead box P1b |
| chr5_-_43959972 | 0.68 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr20_-_25626428 | 0.68 |
ENSDART00000136475
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr6_+_40723554 | 0.68 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr7_-_18545515 | 0.67 |
ENSDART00000040534
|
rgs12a
|
regulator of G protein signaling 12a |
| chr4_-_39111612 | 0.67 |
ENSDART00000150394
|
si:dkey-122c11.8
|
si:dkey-122c11.8 |
| chr20_-_47347962 | 0.66 |
ENSDART00000080863
|
dtnba
|
dystrobrevin, beta a |
| chr20_+_30578967 | 0.66 |
ENSDART00000010494
|
fgfr1op
|
FGFR1 oncogene partner |
| chr17_+_15413412 | 0.66 |
ENSDART00000149871
|
cx40.8
|
connexin 40.8 |
| chr23_+_14217508 | 0.65 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr11_-_9948487 | 0.65 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr12_+_46967789 | 0.64 |
ENSDART00000114866
|
oat
|
ornithine aminotransferase |
| chr20_+_12830448 | 0.64 |
ENSDART00000164754
|
BX547930.4
|
|
| chr17_+_53424415 | 0.63 |
ENSDART00000157022
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr1_-_13989643 | 0.63 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr9_+_32930622 | 0.63 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr21_-_41839683 | 0.62 |
ENSDART00000160908
ENSDART00000137630 |
cct6a
|
chaperonin containing TCP1, subunit 6A (zeta 1) |
| chr15_-_4967302 | 0.62 |
ENSDART00000101992
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr10_-_4375190 | 0.62 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr21_+_5993188 | 0.62 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr24_-_12983829 | 0.61 |
ENSDART00000133324
|
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr13_-_40499296 | 0.61 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr6_+_18441331 | 0.61 |
ENSDART00000171005
|
si:dkey-31g6.4
|
si:dkey-31g6.4 |
| chr15_-_4967490 | 0.61 |
ENSDART00000180551
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 1.3 | 3.9 | GO:0045141 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.7 | 10.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.4 | 1.2 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.4 | 1.2 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 1.0 | GO:1903961 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.3 | 1.3 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 1.1 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.3 | 1.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 2.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.3 | 1.3 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.2 | 0.7 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.2 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.7 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.7 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.2 | 1.6 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 2.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.9 | GO:0001778 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.2 | 0.8 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.2 | 1.2 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 1.0 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.2 | 1.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.2 | 0.5 | GO:0090247 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.2 | 1.6 | GO:1904950 | negative regulation of protein transport(GO:0051224) negative regulation of establishment of protein localization(GO:1904950) |
| 0.2 | 0.6 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.2 | 1.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 2.9 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.6 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.6 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 1.6 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 1.5 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 2.4 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.5 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.4 | GO:0005984 | disaccharide metabolic process(GO:0005984) sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.9 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.8 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.7 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.1 | 1.6 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 0.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.4 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.6 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.6 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 1.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.3 | GO:0045023 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.6 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 1.0 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.2 | GO:0048389 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.6 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.9 | GO:0001964 | startle response(GO:0001964) |
| 0.1 | 0.9 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 1.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.5 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.2 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.1 | 0.6 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.6 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.9 | GO:0035194 | posttranscriptional gene silencing(GO:0016441) posttranscriptional gene silencing by RNA(GO:0035194) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.6 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.5 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.8 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 1.0 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.7 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 2.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.6 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.2 | GO:0007618 | mating(GO:0007618) |
| 0.0 | 1.1 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.5 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 1.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 3.5 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 1.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 1.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 4.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.7 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 2.4 | GO:0048477 | oogenesis(GO:0048477) |
| 0.0 | 0.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 1.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 10.6 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.7 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.9 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 1.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.3 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0033153 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.9 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.4 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 2.1 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.7 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.6 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.0 | 0.2 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.2 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.9 | GO:0006612 | protein targeting to membrane(GO:0006612) |
| 0.0 | 0.2 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.5 | 5.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.4 | 4.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.4 | 2.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.3 | 4.4 | GO:0070187 | telosome(GO:0070187) |
| 0.3 | 2.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 1.1 | GO:0097433 | dense body(GO:0097433) |
| 0.2 | 1.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.9 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 2.9 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 2.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.5 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 1.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 3.5 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 1.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.5 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 3.2 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 2.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.9 | 5.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.6 | 3.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.5 | 2.8 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.4 | 1.3 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.4 | 2.6 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.4 | 4.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.4 | 1.2 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.4 | 1.6 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.4 | 1.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.4 | 2.8 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 1.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 1.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 1.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.7 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 1.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.2 | 2.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.7 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 1.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.2 | 0.8 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.2 | 2.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.2 | 0.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 1.1 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 0.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 2.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 1.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.7 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 1.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.4 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.1 | 1.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 2.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.6 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.5 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 2.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.7 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.7 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 3.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.4 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.6 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.6 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 3.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 2.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.2 | GO:0005231 | excitatory extracellular ligand-gated ion channel activity(GO:0005231) |
| 0.0 | 0.8 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 5.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.9 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.2 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 2.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.3 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 2.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 1.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 3.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.9 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |