Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
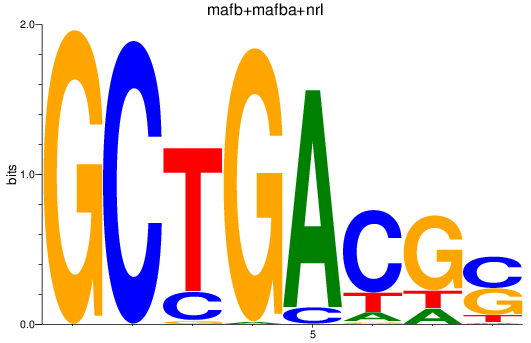
Results for mafb+mafba+nrl
Z-value: 0.85
Transcription factors associated with mafb+mafba+nrl
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafba
|
ENSDARG00000017121 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ba |
|
mafb
|
ENSDARG00000076520 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog b (paralog b) |
|
nrl
|
ENSDARG00000100466 | neural retina leucine zipper |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafb | dr11_v1_chr25_+_36674715_36674715 | 0.63 | 4.9e-03 | Click! |
| nrl | dr11_v1_chr20_+_661037_661037 | -0.27 | 2.8e-01 | Click! |
| mafba | dr11_v1_chr23_-_3408777_3408777 | -0.10 | 7.1e-01 | Click! |
Activity profile of mafb+mafba+nrl motif
Sorted Z-values of mafb+mafba+nrl motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_4482449 | 1.72 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr24_+_13277573 | 1.60 |
ENSDART00000137886
|
si:ch211-171b20.3
|
si:ch211-171b20.3 |
| chr5_-_71722257 | 1.42 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr16_-_54465271 | 1.16 |
ENSDART00000185978
|
cyp11c1
|
cytochrome P450, family 11, subfamily C, polypeptide 1 |
| chr5_+_62723233 | 1.08 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr14_-_24397718 | 1.04 |
ENSDART00000110164
|
si:ch211-163m17.4
|
si:ch211-163m17.4 |
| chr19_+_233143 | 1.01 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr13_+_50375800 | 1.01 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr25_-_19584735 | 0.91 |
ENSDART00000137930
|
sycp3
|
synaptonemal complex protein 3 |
| chr1_-_59287410 | 0.90 |
ENSDART00000158011
ENSDART00000170580 |
col5a3b
|
collagen, type V, alpha 3b |
| chr25_+_245438 | 0.87 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr12_-_27242498 | 0.86 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr4_+_15819728 | 0.86 |
ENSDART00000101613
|
lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr15_-_106689 | 0.86 |
ENSDART00000185007
|
apoa1b
|
apolipoprotein A-Ib |
| chr6_-_39313027 | 0.84 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr19_+_9004256 | 0.79 |
ENSDART00000186401
ENSDART00000091443 |
si:ch211-81a5.1
|
si:ch211-81a5.1 |
| chr11_+_39135050 | 0.77 |
ENSDART00000180571
ENSDART00000189685 |
cdc42
|
cell division cycle 42 |
| chr6_+_120181 | 0.76 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr2_-_14798295 | 0.74 |
ENSDART00000143430
ENSDART00000145869 |
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr19_-_5332784 | 0.74 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chrM_+_3803 | 0.71 |
ENSDART00000093596
|
mt-nd1
|
NADH dehydrogenase 1, mitochondrial |
| chr16_-_42969598 | 0.71 |
ENSDART00000156011
|
si:ch211-135n15.3
|
si:ch211-135n15.3 |
| chr5_+_42467867 | 0.70 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr3_+_1209006 | 0.70 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr23_-_45049044 | 0.67 |
ENSDART00000067630
|
SH3GL2 (1 of many)
|
SH3 domain containing GRB2 like 2, endophilin A1 |
| chr23_+_6795531 | 0.66 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr15_-_19771981 | 0.65 |
ENSDART00000175502
ENSDART00000159475 |
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr22_+_18816662 | 0.64 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr21_+_40106448 | 0.61 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_-_4023050 | 0.60 |
ENSDART00000162883
|
stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr20_-_53981626 | 0.60 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr24_-_35707552 | 0.60 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr4_-_12795030 | 0.59 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr21_+_26726936 | 0.59 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr9_-_105135 | 0.59 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr14_+_33329420 | 0.58 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr17_-_39786222 | 0.57 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr18_+_7611298 | 0.57 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr5_+_42402536 | 0.57 |
ENSDART00000186754
|
BX548073.11
|
|
| chr23_+_32011768 | 0.57 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr23_+_44644911 | 0.55 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr5_-_67661102 | 0.55 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr13_+_24755049 | 0.55 |
ENSDART00000134102
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr2_-_10450784 | 0.54 |
ENSDART00000081196
|
scinlb
|
scinderin like b |
| chr8_+_53311965 | 0.54 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr5_-_33259079 | 0.54 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr17_+_14425219 | 0.53 |
ENSDART00000153994
|
ccdc175
|
coiled-coil domain containing 175 |
| chr21_+_244503 | 0.53 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr22_-_193234 | 0.52 |
ENSDART00000131067
|
fbxo42
|
F-box protein 42 |
| chr17_+_46739693 | 0.52 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr11_+_45347961 | 0.51 |
ENSDART00000172962
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr25_+_28823952 | 0.51 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr4_-_12795436 | 0.50 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr19_+_58954 | 0.49 |
ENSDART00000162379
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr4_+_38344 | 0.49 |
ENSDART00000170197
ENSDART00000175348 |
phtf2
|
putative homeodomain transcription factor 2 |
| chr23_+_6795709 | 0.49 |
ENSDART00000149136
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr23_+_2542158 | 0.49 |
ENSDART00000182197
|
LO017835.1
|
|
| chr18_+_30028135 | 0.49 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr16_+_1254390 | 0.48 |
ENSDART00000092627
|
ADAMTSL4
|
ADAMTS like 4 |
| chr5_-_68022631 | 0.48 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr2_+_35728033 | 0.47 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr9_-_42696408 | 0.47 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr25_-_27665978 | 0.46 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr17_+_10593398 | 0.45 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr5_+_20112032 | 0.45 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr10_-_26744131 | 0.45 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr4_-_890220 | 0.45 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr25_-_22187397 | 0.44 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr1_+_22003173 | 0.44 |
ENSDART00000054388
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr16_+_54588930 | 0.44 |
ENSDART00000159174
|
dennd4b
|
DENN/MADD domain containing 4B |
| chr20_-_1383916 | 0.44 |
ENSDART00000152373
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr22_+_22021936 | 0.44 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr8_+_19548985 | 0.44 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr4_-_73190246 | 0.43 |
ENSDART00000170842
|
LO018260.1
|
|
| chr11_+_26669724 | 0.43 |
ENSDART00000157157
|
si:dkey-57n24.6
|
si:dkey-57n24.6 |
| chr5_-_56119028 | 0.43 |
ENSDART00000083134
|
mks1
|
Meckel syndrome, type 1 |
| chr24_+_38522254 | 0.42 |
ENSDART00000156189
|
si:ch1073-66l23.1
|
si:ch1073-66l23.1 |
| chr22_-_1079773 | 0.42 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr16_-_42894628 | 0.42 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr15_-_28095532 | 0.42 |
ENSDART00000191490
|
cryba1a
|
crystallin, beta A1a |
| chr25_-_3627130 | 0.41 |
ENSDART00000171863
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr3_+_40809892 | 0.41 |
ENSDART00000190803
|
arpc1b
|
actin related protein 2/3 complex, subunit 1B |
| chr17_+_51940768 | 0.41 |
ENSDART00000053422
|
ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr19_-_10081047 | 0.40 |
ENSDART00000128837
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr1_+_11711226 | 0.40 |
ENSDART00000147623
|
si:dkey-26i13.8
|
si:dkey-26i13.8 |
| chr3_+_55100045 | 0.40 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr16_+_19014886 | 0.40 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr18_-_48755037 | 0.40 |
ENSDART00000109673
|
tirap
|
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr25_-_16600811 | 0.40 |
ENSDART00000011397
|
cpa2
|
carboxypeptidase A2 (pancreatic) |
| chr7_-_33130552 | 0.40 |
ENSDART00000127006
|
mns1
|
meiosis-specific nuclear structural 1 |
| chr9_-_24985511 | 0.39 |
ENSDART00000141957
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr10_+_10351685 | 0.39 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr25_+_16601839 | 0.39 |
ENSDART00000008986
|
atp6v1e1a
|
ATPase H+ transporting V1 subunit E1a |
| chr15_+_46386261 | 0.39 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr4_-_191736 | 0.39 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr2_+_31957554 | 0.39 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr6_+_7250824 | 0.39 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr1_-_11878845 | 0.38 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr16_-_29194517 | 0.38 |
ENSDART00000046114
ENSDART00000148899 |
mef2d
|
myocyte enhancer factor 2d |
| chr15_+_2559875 | 0.38 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr21_-_45920 | 0.38 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr8_+_53464216 | 0.38 |
ENSDART00000169514
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr16_-_2558653 | 0.38 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr23_+_7710447 | 0.37 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr4_-_8030583 | 0.37 |
ENSDART00000113628
|
si:ch211-240l19.8
|
si:ch211-240l19.8 |
| chr12_-_49151326 | 0.37 |
ENSDART00000153244
|
bub3
|
BUB3 mitotic checkpoint protein |
| chr18_-_20869175 | 0.37 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr15_+_3284684 | 0.37 |
ENSDART00000179778
|
foxo1a
|
forkhead box O1 a |
| chr13_-_38792061 | 0.37 |
ENSDART00000162858
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr7_+_39166460 | 0.36 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr15_-_45246911 | 0.36 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr6_+_24661736 | 0.36 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr3_+_55105066 | 0.36 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr16_+_10841163 | 0.36 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr12_+_466324 | 0.36 |
ENSDART00000169691
|
mprip
|
myosin phosphatase Rho interacting protein |
| chr13_+_22675802 | 0.36 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr1_-_59407322 | 0.36 |
ENSDART00000161661
|
si:ch211-188p14.5
|
si:ch211-188p14.5 |
| chr8_-_15305528 | 0.35 |
ENSDART00000134794
|
spata6
|
spermatogenesis associated 6 |
| chr1_-_59232267 | 0.35 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr23_+_7710721 | 0.35 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr14_+_23518110 | 0.34 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr7_-_25697285 | 0.34 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr8_-_43456025 | 0.34 |
ENSDART00000001092
ENSDART00000140618 |
ncor2
|
nuclear receptor corepressor 2 |
| chr12_-_684200 | 0.34 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr4_-_149334 | 0.34 |
ENSDART00000163280
|
tbk1
|
TANK-binding kinase 1 |
| chr25_-_36361697 | 0.34 |
ENSDART00000152388
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr18_+_14477740 | 0.34 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr19_+_23982466 | 0.34 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr14_+_49135264 | 0.34 |
ENSDART00000084119
|
si:ch1073-44g3.1
|
si:ch1073-44g3.1 |
| chr23_-_26077038 | 0.34 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr23_+_35713557 | 0.34 |
ENSDART00000123518
|
tuba1c
|
tubulin, alpha 1c |
| chr14_+_81919 | 0.34 |
ENSDART00000171444
|
stag3
|
stromal antigen 3 |
| chr12_-_3840664 | 0.33 |
ENSDART00000160967
|
taok2b
|
TAO kinase 2b |
| chr8_+_52314542 | 0.33 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr23_-_45318760 | 0.33 |
ENSDART00000166883
|
ccdc171
|
coiled-coil domain containing 171 |
| chr22_+_38192568 | 0.33 |
ENSDART00000126323
|
cp
|
ceruloplasmin |
| chr3_+_32425202 | 0.33 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr19_+_56351 | 0.33 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr16_+_11242443 | 0.33 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr1_+_54606354 | 0.33 |
ENSDART00000125227
|
si:ch211-202h22.10
|
si:ch211-202h22.10 |
| chr16_+_23282655 | 0.33 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr21_+_30502002 | 0.32 |
ENSDART00000043727
|
slc7a3a
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3a |
| chr15_-_19772372 | 0.32 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr9_-_5916693 | 0.32 |
ENSDART00000115404
ENSDART00000148792 |
GPR89A
|
si:ch73-390b10.2 |
| chr19_-_6134802 | 0.32 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr7_-_12968689 | 0.31 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr1_-_11973341 | 0.31 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr15_-_28094256 | 0.31 |
ENSDART00000142041
ENSDART00000132153 ENSDART00000146657 ENSDART00000048720 |
cryba1a
|
crystallin, beta A1a |
| chr1_-_55183088 | 0.31 |
ENSDART00000100619
|
LUC7L2
|
zgc:158803 |
| chr22_+_38301365 | 0.31 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr19_-_379750 | 0.31 |
ENSDART00000102725
|
tnfaip8l2a
|
tumor necrosis factor, alpha-induced protein 8-like 2a |
| chr1_+_10318089 | 0.31 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr24_+_3478871 | 0.31 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr3_+_20012891 | 0.31 |
ENSDART00000078974
|
tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr4_-_760560 | 0.31 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr23_+_44157682 | 0.30 |
ENSDART00000164474
ENSDART00000149928 |
si:ch73-106g13.1
|
si:ch73-106g13.1 |
| chr8_-_25074319 | 0.30 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr16_+_6021908 | 0.30 |
ENSDART00000163786
|
BX511115.1
|
|
| chr14_+_33329761 | 0.30 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr11_+_30161168 | 0.29 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr21_-_43015383 | 0.29 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr23_+_25210953 | 0.29 |
ENSDART00000132799
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr19_+_156757 | 0.29 |
ENSDART00000167717
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr24_-_12938922 | 0.29 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr9_-_296169 | 0.29 |
ENSDART00000165228
|
kif5aa
|
kinesin family member 5A, a |
| chr10_-_42147318 | 0.28 |
ENSDART00000134890
|
dusp11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr17_-_30521043 | 0.28 |
ENSDART00000087111
|
itsn2b
|
intersectin 2b |
| chr10_+_31809226 | 0.28 |
ENSDART00000087898
|
foxo1b
|
forkhead box O1 b |
| chr15_+_3284416 | 0.28 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr21_-_20932603 | 0.27 |
ENSDART00000138155
ENSDART00000079709 |
c6
|
complement component 6 |
| chr14_-_32089117 | 0.27 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr25_-_3503164 | 0.27 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr25_+_19999623 | 0.27 |
ENSDART00000026401
|
zgc:194665
|
zgc:194665 |
| chr10_+_25355308 | 0.27 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr18_-_12451772 | 0.27 |
ENSDART00000175083
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr15_+_15479442 | 0.27 |
ENSDART00000067095
|
nek8
|
NIMA-related kinase 8 |
| chr19_-_15192638 | 0.27 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr23_+_23658474 | 0.27 |
ENSDART00000162838
|
agrn
|
agrin |
| chr13_+_233482 | 0.27 |
ENSDART00000102511
|
cfap36
|
cilia and flagella associated protein 36 |
| chr23_-_28141419 | 0.27 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr20_+_35086242 | 0.27 |
ENSDART00000114262
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr22_+_28969071 | 0.26 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr1_+_10129099 | 0.26 |
ENSDART00000187740
|
rbm46
|
RNA binding motif protein 46 |
| chr20_+_39685572 | 0.26 |
ENSDART00000050729
|
hddc2
|
HD domain containing 2 |
| chr7_-_73848458 | 0.26 |
ENSDART00000041385
|
zgc:163061
|
zgc:163061 |
| chr3_-_46410387 | 0.26 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr7_-_26087807 | 0.26 |
ENSDART00000052989
|
ache
|
acetylcholinesterase |
| chr25_-_3503458 | 0.26 |
ENSDART00000173269
|
PRKAR2B
|
si:ch211-272n13.7 |
| chr4_-_4592287 | 0.26 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr23_-_270847 | 0.26 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr8_-_25011157 | 0.26 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr22_-_7669624 | 0.25 |
ENSDART00000189617
|
AL929276.2
|
|
| chr21_+_35215810 | 0.25 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr21_-_22543611 | 0.25 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr7_-_29200090 | 0.25 |
ENSDART00000173973
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr2_-_44720551 | 0.25 |
ENSDART00000146380
|
map6d1
|
MAP6 domain containing 1 |
| chr4_+_16710001 | 0.25 |
ENSDART00000035899
|
pkp2
|
plakophilin 2 |
| chr21_+_382400 | 0.25 |
ENSDART00000190785
ENSDART00000124615 |
rad23b
|
RAD23 homolog B, nucleotide excision repair protein |
| chr6_-_8311044 | 0.25 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr2_+_56139941 | 0.25 |
ENSDART00000164741
|
pgpep1
|
pyroglutamyl-peptidase I |
Network of associatons between targets according to the STRING database.
First level regulatory network of mafb+mafba+nrl
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 1.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.7 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 0.9 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.5 | GO:0035376 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.1 | 0.5 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.1 | 0.4 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.9 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.1 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.3 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.6 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 0.7 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.8 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.3 | GO:0042117 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.2 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 1.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.3 | GO:0044003 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 | 0.3 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.3 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 0.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.1 | 0.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.3 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 0.2 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 0.3 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.6 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.3 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.2 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.3 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.1 | 0.6 | GO:1902110 | positive regulation of mitochondrial membrane permeability(GO:0035794) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.1 | 0.2 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.3 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.1 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.1 | 0.5 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.2 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.3 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.2 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.1 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.2 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.2 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.2 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.0 | 1.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.5 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.0 | 0.5 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.4 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.4 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.2 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.2 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.6 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0046902 | regulation of mitochondrial membrane permeability(GO:0046902) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.5 | GO:0031629 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.4 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.1 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.2 | GO:0060211 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.0 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 0.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.3 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.0 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.1 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.0 | GO:0090183 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.6 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 1.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.9 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.9 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.4 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.3 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.1 | 0.3 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.3 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 0.2 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.5 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.9 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0042974 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0022884 | macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.8 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 1.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.6 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |