Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
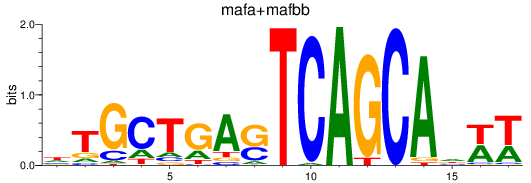
Results for mafa+mafbb
Z-value: 0.47
Transcription factors associated with mafa+mafbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
mafa
|
ENSDARG00000015890 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
|
mafbb
|
ENSDARG00000070542 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
|
mafbb
|
ENSDARG00000099120 | v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mafa | dr11_v1_chr18_+_29402623_29402623 | 0.89 | 5.8e-07 | Click! |
| mafbb | dr11_v1_chr11_-_25384213_25384213 | -0.02 | 9.3e-01 | Click! |
Activity profile of mafa+mafbb motif
Sorted Z-values of mafa+mafbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_31222433 | 2.56 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr3_-_18030938 | 2.37 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr22_-_25469751 | 1.55 |
ENSDART00000171670
|
CR769772.4
|
|
| chr18_+_7456597 | 1.19 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr24_+_22022109 | 1.19 |
ENSDART00000133686
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr24_+_22021675 | 1.06 |
ENSDART00000081234
|
ropn1l
|
rhophilin associated tail protein 1-like |
| chr13_+_2908764 | 1.03 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr2_-_24554416 | 0.99 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr8_+_31435452 | 0.96 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr25_-_7764083 | 0.86 |
ENSDART00000179800
ENSDART00000181858 |
phf21ab
|
PHD finger protein 21Ab |
| chr18_+_22302635 | 0.84 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr13_-_37642890 | 0.81 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr4_+_9609905 | 0.78 |
ENSDART00000142284
ENSDART00000150687 |
meig1
|
meiosis/spermiogenesis associated 1 |
| chr1_+_49652967 | 0.76 |
ENSDART00000191296
|
tsga10
|
testis specific, 10 |
| chr6_-_40744720 | 0.75 |
ENSDART00000154916
ENSDART00000186922 |
p4htm
|
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr21_-_5393125 | 0.73 |
ENSDART00000146061
|
psmd5
|
proteasome 26S subunit, non-ATPase 5 |
| chr21_+_19347655 | 0.71 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr10_+_26747755 | 0.71 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr16_+_7662609 | 0.71 |
ENSDART00000184895
ENSDART00000149404 ENSDART00000081418 ENSDART00000081422 |
bves
|
blood vessel epicardial substance |
| chr13_-_23007813 | 0.69 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr1_-_52498146 | 0.68 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr22_-_24285432 | 0.68 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr11_-_40128722 | 0.67 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr15_-_47822597 | 0.67 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr2_+_17525355 | 0.59 |
ENSDART00000074101
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr14_-_26397849 | 0.54 |
ENSDART00000148140
|
pimr212
|
Pim proto-oncogene, serine/threonine kinase, related 212 |
| chr6_+_33885828 | 0.53 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr19_+_1465004 | 0.53 |
ENSDART00000159157
|
CABZ01073736.1
|
|
| chr23_-_26077038 | 0.52 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr16_+_46294337 | 0.50 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr19_-_35428815 | 0.49 |
ENSDART00000169006
ENSDART00000003167 |
ak2
|
adenylate kinase 2 |
| chr11_-_5865744 | 0.49 |
ENSDART00000104360
|
gamt
|
guanidinoacetate N-methyltransferase |
| chr7_+_35068036 | 0.49 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr7_-_25697285 | 0.48 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr17_-_19345521 | 0.48 |
ENSDART00000082085
|
gsc
|
goosecoid |
| chr1_-_52497834 | 0.45 |
ENSDART00000136469
ENSDART00000004233 |
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_-_49859747 | 0.45 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr17_+_6217704 | 0.45 |
ENSDART00000129100
|
BX000363.1
|
|
| chr10_+_28428222 | 0.43 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr10_+_10801564 | 0.43 |
ENSDART00000027026
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr16_-_49781481 | 0.42 |
ENSDART00000127513
|
znf385d
|
zinc finger protein 385D |
| chr7_+_38811800 | 0.42 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr9_-_2573121 | 0.42 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr20_+_812012 | 0.42 |
ENSDART00000179299
ENSDART00000137135 |
mei4
|
meiosis-specific, MEI4 homolog (S. cerevisiae) |
| chr23_+_7379728 | 0.41 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr9_+_14291932 | 0.41 |
ENSDART00000183119
|
BX511082.1
|
|
| chr1_-_10847753 | 0.40 |
ENSDART00000141052
|
dmd
|
dystrophin |
| chr8_-_41228530 | 0.39 |
ENSDART00000165949
ENSDART00000173055 |
fahd2a
|
fumarylacetoacetate hydrolase domain containing 2A |
| chr23_-_20764227 | 0.38 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr2_+_25816843 | 0.38 |
ENSDART00000078639
|
slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr6_+_52247993 | 0.37 |
ENSDART00000172176
|
zc3h10
|
zinc finger CCCH-type containing 10 |
| chr1_-_26131088 | 0.37 |
ENSDART00000193973
ENSDART00000054209 |
cdkn2a/b
|
cyclin-dependent kinase inhibitor 2A/B (p15, inhibits CDK4) |
| chr23_+_42304602 | 0.37 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr20_+_46385907 | 0.36 |
ENSDART00000060710
|
adgrg11
|
adhesion G protein-coupled receptor G11 |
| chr7_-_6415991 | 0.36 |
ENSDART00000173349
|
CU457819.3
|
Histone H3.2 |
| chr23_+_4253957 | 0.35 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr17_+_23938283 | 0.35 |
ENSDART00000184391
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr14_+_146857 | 0.35 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr20_+_18741089 | 0.34 |
ENSDART00000183218
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr22_+_28888781 | 0.34 |
ENSDART00000145629
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr13_-_20381485 | 0.34 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr10_+_10801719 | 0.34 |
ENSDART00000193648
|
ambp
|
alpha-1-microglobulin/bikunin precursor |
| chr21_+_45659525 | 0.33 |
ENSDART00000160391
|
slc22a5
|
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
| chr2_-_9748039 | 0.32 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr9_+_32930622 | 0.32 |
ENSDART00000100928
|
cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr4_+_47257854 | 0.32 |
ENSDART00000173868
|
crestin
|
crestin |
| chr17_-_53353653 | 0.32 |
ENSDART00000180744
ENSDART00000026879 |
unm_sa911
|
un-named sa911 |
| chr22_-_38516922 | 0.31 |
ENSDART00000151257
ENSDART00000151785 |
klc4
|
kinesin light chain 4 |
| chr2_-_9059955 | 0.31 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr11_-_33960318 | 0.31 |
ENSDART00000087597
|
col6a2
|
collagen, type VI, alpha 2 |
| chr10_-_300000 | 0.31 |
ENSDART00000183273
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr22_-_10354381 | 0.31 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr10_+_466926 | 0.30 |
ENSDART00000145220
|
arvcfa
|
ARVCF, delta catenin family member a |
| chr2_-_30611389 | 0.30 |
ENSDART00000142500
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr9_-_23850978 | 0.30 |
ENSDART00000132915
|
serpine3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3 |
| chr23_-_35082494 | 0.29 |
ENSDART00000189809
|
BX294434.1
|
|
| chr24_-_12938922 | 0.28 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr17_+_51262556 | 0.28 |
ENSDART00000186748
ENSDART00000181606 ENSDART00000063738 ENSDART00000189066 |
eipr1
|
EARP complex and GARP complex interacting protein 1 |
| chr8_+_23439340 | 0.28 |
ENSDART00000109932
ENSDART00000185469 |
foxp3b
|
forkhead box P3b |
| chr2_-_3038904 | 0.27 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr9_-_2572790 | 0.27 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr20_-_39219537 | 0.27 |
ENSDART00000005764
|
cyp39a1
|
cytochrome P450, family 39, subfamily A, polypeptide 1 |
| chr1_-_7582859 | 0.26 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr1_-_7970653 | 0.26 |
ENSDART00000188909
|
si:dkey-79f11.7
|
si:dkey-79f11.7 |
| chr9_-_55586151 | 0.25 |
ENSDART00000181886
|
arsh
|
arylsulfatase H |
| chr12_+_16967715 | 0.25 |
ENSDART00000138174
|
slc16a12b
|
solute carrier family 16, member 12b |
| chr2_+_38373272 | 0.25 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr17_-_50422584 | 0.25 |
ENSDART00000157095
|
si:ch211-235i11.5
|
si:ch211-235i11.5 |
| chr22_-_27115241 | 0.25 |
ENSDART00000019442
|
xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr10_+_38708099 | 0.25 |
ENSDART00000172306
|
tmprss2
|
transmembrane protease, serine 2 |
| chr7_+_63325819 | 0.25 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr20_-_47027379 | 0.25 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr19_+_170705 | 0.24 |
ENSDART00000053622
|
rnf139
|
ring finger protein 139 |
| chr23_-_44848961 | 0.24 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr6_+_40723554 | 0.23 |
ENSDART00000103833
|
slc26a6l
|
solute carrier family 26, member 6, like |
| chr9_+_13985567 | 0.23 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr16_-_44349845 | 0.23 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr23_-_16666583 | 0.23 |
ENSDART00000189804
ENSDART00000136496 |
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr21_-_28523548 | 0.23 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr18_+_33725576 | 0.23 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr21_+_45496442 | 0.22 |
ENSDART00000164880
|
si:dkey-223p19.1
|
si:dkey-223p19.1 |
| chr8_+_23436411 | 0.22 |
ENSDART00000140044
|
foxp3b
|
forkhead box P3b |
| chr14_+_34547554 | 0.22 |
ENSDART00000074819
|
gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr24_-_26369185 | 0.21 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr20_+_37300296 | 0.21 |
ENSDART00000180789
|
vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr1_-_12278522 | 0.21 |
ENSDART00000142122
ENSDART00000003825 |
cplx2l
|
complexin 2, like |
| chr6_-_426041 | 0.20 |
ENSDART00000162789
|
fam83fb
|
family with sequence similarity 83, member Fb |
| chr20_-_23946296 | 0.20 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr5_-_43959972 | 0.20 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr15_-_47838391 | 0.20 |
ENSDART00000180337
|
kbtbd3
|
kelch repeat and BTB (POZ) domain containing 3 |
| chr25_-_35045250 | 0.19 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr12_-_3940768 | 0.19 |
ENSDART00000134292
|
zgc:92040
|
zgc:92040 |
| chr20_+_23408970 | 0.19 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr21_-_11820379 | 0.19 |
ENSDART00000126640
|
RHOBTB3
|
si:dkey-6b12.5 |
| chr16_-_12319822 | 0.18 |
ENSDART00000127453
ENSDART00000184526 |
trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr7_+_42460302 | 0.18 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr1_+_54766943 | 0.18 |
ENSDART00000144759
|
nlrc6
|
NLR family CARD domain containing 6 |
| chr7_+_57108823 | 0.18 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr19_+_31771270 | 0.18 |
ENSDART00000147474
|
stmn2b
|
stathmin 2b |
| chr16_+_13855039 | 0.17 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr20_+_12830448 | 0.17 |
ENSDART00000164754
|
BX547930.4
|
|
| chr20_-_20613584 | 0.17 |
ENSDART00000140991
|
si:ch211-253p18.2
|
si:ch211-253p18.2 |
| chr20_+_18661624 | 0.17 |
ENSDART00000152136
ENSDART00000126959 |
tnfaip2a
|
tumor necrosis factor, alpha-induced protein 2a |
| chr20_-_29420713 | 0.16 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr1_-_30457062 | 0.16 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr1_-_54706039 | 0.16 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr7_+_23875269 | 0.15 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr9_+_6802641 | 0.15 |
ENSDART00000187278
|
FO203514.1
|
|
| chr1_+_8601935 | 0.15 |
ENSDART00000152367
|
si:ch211-160d14.6
|
si:ch211-160d14.6 |
| chr17_-_20236228 | 0.14 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr10_-_40514643 | 0.14 |
ENSDART00000140705
|
taar19k
|
trace amine associated receptor 19k |
| chr11_-_36263886 | 0.14 |
ENSDART00000140397
|
nfya
|
nuclear transcription factor Y, alpha |
| chr23_-_44494401 | 0.14 |
ENSDART00000114640
ENSDART00000148532 |
actl6b
|
actin-like 6B |
| chr4_+_77709678 | 0.14 |
ENSDART00000036856
|
AL935186.1
|
|
| chr5_-_30475011 | 0.14 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr13_-_35459928 | 0.14 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr23_-_19500559 | 0.13 |
ENSDART00000177414
ENSDART00000145898 |
asb14b
|
ankyrin repeat and SOCS box containing 14b |
| chr13_+_36770738 | 0.13 |
ENSDART00000146696
|
atl1
|
atlastin GTPase 1 |
| chr15_-_29388012 | 0.13 |
ENSDART00000115032
|
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr16_-_31435020 | 0.13 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr19_+_7453996 | 0.13 |
ENSDART00000147491
ENSDART00000136502 |
mindy1
|
MINDY lysine 48 deubiquitinase 1 |
| chr8_-_50287949 | 0.13 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr20_+_23315242 | 0.12 |
ENSDART00000132248
|
fryl
|
furry homolog, like |
| chr13_+_29773153 | 0.11 |
ENSDART00000144443
ENSDART00000133796 ENSDART00000141310 ENSDART00000139782 |
pax2a
|
paired box 2a |
| chr15_-_29387446 | 0.11 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr7_+_27317174 | 0.11 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr6_-_13783604 | 0.11 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr15_+_3790556 | 0.11 |
ENSDART00000181467
|
RNF14 (1 of many)
|
ring finger protein 14 |
| chr20_-_20410029 | 0.11 |
ENSDART00000192177
ENSDART00000063483 |
prkchb
|
protein kinase C, eta, b |
| chr9_+_38372216 | 0.10 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr11_+_42765963 | 0.10 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr6_+_58569808 | 0.10 |
ENSDART00000154393
|
helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr8_-_53926228 | 0.09 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr17_-_31044803 | 0.09 |
ENSDART00000185941
ENSDART00000152016 |
eml1
|
echinoderm microtubule associated protein like 1 |
| chr25_+_336503 | 0.09 |
ENSDART00000160395
|
CU929262.1
|
|
| chr9_-_8670158 | 0.09 |
ENSDART00000077296
|
col4a1
|
collagen, type IV, alpha 1 |
| chr4_+_68562464 | 0.09 |
ENSDART00000192954
|
BX548011.4
|
|
| chr23_+_24124684 | 0.09 |
ENSDART00000144478
|
si:dkey-21o19.2
|
si:dkey-21o19.2 |
| chr1_-_30510839 | 0.08 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr8_-_7847921 | 0.07 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr7_+_50053339 | 0.07 |
ENSDART00000174308
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr19_-_9711472 | 0.07 |
ENSDART00000016197
ENSDART00000175075 |
slc2a3a
|
solute carrier family 2 (facilitated glucose transporter), member 3a |
| chr22_-_16416882 | 0.07 |
ENSDART00000062749
|
cts12
|
cathepsin 12 |
| chr25_+_31277415 | 0.07 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr6_-_48087152 | 0.07 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr3_+_634682 | 0.07 |
ENSDART00000164305
|
dicp1.1
|
diverse immunoglobulin domain-containing protein 1.1 |
| chr14_-_4076480 | 0.07 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr19_+_7567763 | 0.07 |
ENSDART00000140411
|
s100a11
|
S100 calcium binding protein A11 |
| chr18_-_45617146 | 0.06 |
ENSDART00000146543
|
wt1b
|
wilms tumor 1b |
| chr5_+_9377005 | 0.06 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr5_-_30615901 | 0.05 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr16_+_31542645 | 0.05 |
ENSDART00000163724
|
SLA (1 of many)
|
Src like adaptor |
| chr5_+_19413838 | 0.05 |
ENSDART00000089014
ENSDART00000146215 |
myo1ha
|
myosin IHa |
| chr14_+_34780886 | 0.05 |
ENSDART00000130469
ENSDART00000188806 ENSDART00000172924 ENSDART00000173266 ENSDART00000166377 ENSDART00000173371 |
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr1_+_50613868 | 0.05 |
ENSDART00000111114
|
HERC5
|
si:ch73-190m4.1 |
| chr15_-_41762530 | 0.05 |
ENSDART00000187125
ENSDART00000154971 |
ftr91
|
finTRIM family, member 91 |
| chr22_-_4892937 | 0.04 |
ENSDART00000147002
|
si:ch73-256j6.4
|
si:ch73-256j6.4 |
| chr14_-_24001825 | 0.04 |
ENSDART00000130704
|
il4
|
interleukin 4 |
| chr17_-_2817035 | 0.04 |
ENSDART00000152002
|
gpr65
|
G protein-coupled receptor 65 |
| chr7_+_31319876 | 0.04 |
ENSDART00000187611
|
fam189a1
|
family with sequence similarity 189, member A1 |
| chr20_-_23852174 | 0.04 |
ENSDART00000122414
|
si:dkey-15j16.6
|
si:dkey-15j16.6 |
| chr24_+_41940299 | 0.04 |
ENSDART00000022349
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr16_-_30564319 | 0.03 |
ENSDART00000145087
|
lmna
|
lamin A |
| chr11_-_23501467 | 0.03 |
ENSDART00000169066
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr10_-_39011514 | 0.03 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr13_-_33153889 | 0.03 |
ENSDART00000141451
|
TC2N
|
tandem C2 domains, nuclear |
| chr16_-_51299061 | 0.03 |
ENSDART00000148677
|
serpinb1l4
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 4 |
| chr11_+_25010491 | 0.03 |
ENSDART00000167285
|
zgc:92107
|
zgc:92107 |
| chr23_-_29376859 | 0.03 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr5_+_38684651 | 0.02 |
ENSDART00000137852
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr11_+_23265157 | 0.02 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr17_-_53359028 | 0.02 |
ENSDART00000185218
|
CABZ01068208.1
|
|
| chr8_+_28629741 | 0.02 |
ENSDART00000163915
|
si:dkey-109a10.2
|
si:dkey-109a10.2 |
| chr6_-_29515709 | 0.02 |
ENSDART00000180205
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr4_+_5317483 | 0.02 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr15_+_1116664 | 0.02 |
ENSDART00000154780
|
p2ry12
|
purinergic receptor P2Y12 |
| chr7_+_39386982 | 0.02 |
ENSDART00000146702
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr24_+_1023839 | 0.02 |
ENSDART00000082526
|
zgc:111976
|
zgc:111976 |
| chr5_+_38685089 | 0.01 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr3_+_709393 | 0.01 |
ENSDART00000191970
|
AL929520.3
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of mafa+mafbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0090220 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.3 | 2.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.2 | 0.5 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.2 | 1.0 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.7 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.1 | 2.5 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.5 | GO:0001778 | plasma membrane repair(GO:0001778) monocyte activation(GO:0042117) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.4 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.3 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.5 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.8 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.3 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 0.7 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0070316 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.9 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 1.0 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.5 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.8 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.0 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.3 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 1.0 | GO:0008430 | selenium binding(GO:0008430) |
| 0.2 | 0.5 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 0.7 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.7 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.6 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.1 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.5 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |