Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
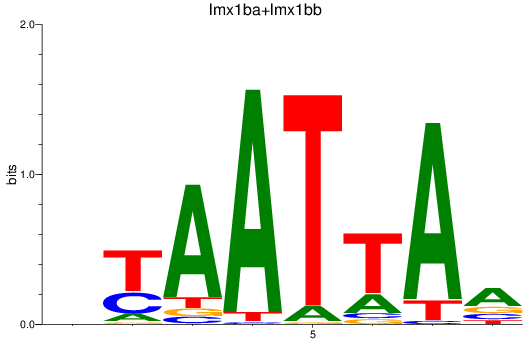
Results for lmx1ba+lmx1bb_lmx1al
Z-value: 0.51
Transcription factors associated with lmx1ba+lmx1bb_lmx1al
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lmx1bb
|
ENSDARG00000068365 | LIM homeobox transcription factor 1, beta b |
|
lmx1ba
|
ENSDARG00000104815 | LIM homeobox transcription factor 1, beta a |
|
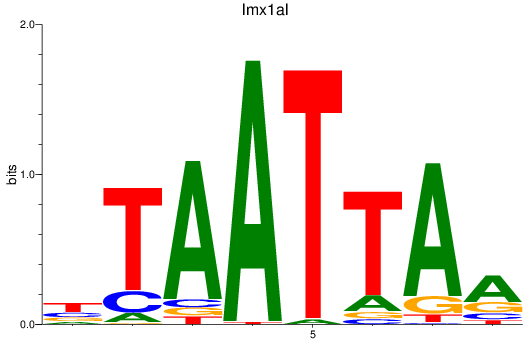
lmx1al
|
ENSDARG00000077915 | LIM homeobox transcription factor 1, alpha-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lmx1al | dr11_v1_chr3_+_52806347_52806441 | 0.83 | 2.2e-05 | Click! |
| lmx1bb | dr11_v1_chr8_-_33381430_33381430 | 0.41 | 8.8e-02 | Click! |
| lmx1ba | dr11_v1_chr5_-_5035683_5035683 | 0.25 | 3.1e-01 | Click! |
Activity profile of lmx1ba+lmx1bb_lmx1al motif
Sorted Z-values of lmx1ba+lmx1bb_lmx1al motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_6954162 | 1.46 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr16_-_42056137 | 1.35 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr20_-_23426339 | 1.01 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_+_40860320 | 0.96 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr22_-_547748 | 0.95 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr23_+_31913292 | 0.93 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr10_-_43771447 | 0.88 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr11_-_41853874 | 0.83 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr23_+_39695827 | 0.83 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr4_+_72723304 | 0.78 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr18_+_19456648 | 0.78 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr16_+_33902006 | 0.76 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr22_+_38037530 | 0.69 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr24_-_35561672 | 0.65 |
ENSDART00000058564
|
mcm4
|
minichromosome maintenance complex component 4 |
| chr15_+_34988148 | 0.64 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr18_-_19456269 | 0.64 |
ENSDART00000060363
|
rpl4
|
ribosomal protein L4 |
| chr20_-_52338782 | 0.64 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr22_-_2937503 | 0.63 |
ENSDART00000092991
ENSDART00000131110 |
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr1_-_18811517 | 0.61 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr3_+_17537352 | 0.61 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr18_+_3037998 | 0.58 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr4_+_22480169 | 0.58 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr14_-_2933185 | 0.56 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr13_+_4225173 | 0.55 |
ENSDART00000058242
ENSDART00000143456 |
mea1
|
male-enhanced antigen 1 |
| chr11_-_38533505 | 0.54 |
ENSDART00000113894
|
slc45a3
|
solute carrier family 45, member 3 |
| chr10_-_26512993 | 0.54 |
ENSDART00000188549
ENSDART00000193316 |
si:dkey-5g14.1
|
si:dkey-5g14.1 |
| chr1_-_46875493 | 0.51 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr20_+_52554352 | 0.51 |
ENSDART00000153217
ENSDART00000145230 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr7_+_72003301 | 0.50 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr2_+_16482338 | 0.50 |
ENSDART00000143912
|
fbxo36b
|
F-box protein 36b |
| chr4_+_9467049 | 0.49 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr10_-_15340362 | 0.47 |
ENSDART00000148119
ENSDART00000127277 ENSDART00000154037 ENSDART00000189109 |
pum3
|
pumilio RNA-binding family member 3 |
| chr24_+_12989727 | 0.47 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr12_-_35830625 | 0.46 |
ENSDART00000180028
|
CU459056.1
|
|
| chr3_+_27798094 | 0.46 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr15_-_4528326 | 0.44 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr9_-_1702648 | 0.43 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr1_-_13989643 | 0.43 |
ENSDART00000191046
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr24_-_38110779 | 0.42 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr14_+_33329420 | 0.42 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr5_+_36850650 | 0.42 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr2_+_50608099 | 0.42 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr19_+_43297546 | 0.41 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr5_+_57210237 | 0.40 |
ENSDART00000167660
|
pja2
|
praja ring finger ubiquitin ligase 2 |
| chr19_-_13808630 | 0.38 |
ENSDART00000166895
ENSDART00000187670 |
ctgfb
|
connective tissue growth factor b |
| chr23_+_355021 | 0.38 |
ENSDART00000190738
|
ap4b1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr7_+_7048245 | 0.36 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr25_+_19739665 | 0.36 |
ENSDART00000067353
|
zgc:101783
|
zgc:101783 |
| chr9_-_20372977 | 0.36 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr17_-_13007484 | 0.36 |
ENSDART00000156812
|
si:dkeyp-33b5.4
|
si:dkeyp-33b5.4 |
| chr1_+_52929185 | 0.36 |
ENSDART00000147683
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr11_-_41220794 | 0.35 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr10_-_8053753 | 0.35 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr20_+_7584211 | 0.34 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr25_-_3867990 | 0.34 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr11_+_17984354 | 0.33 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr19_+_45962016 | 0.33 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr24_-_37698796 | 0.32 |
ENSDART00000172178
|
si:ch211-231f6.6
|
si:ch211-231f6.6 |
| chr13_+_31402067 | 0.32 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| chr17_+_15433518 | 0.32 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr23_+_31912882 | 0.32 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr20_-_31075972 | 0.32 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr3_-_16719244 | 0.31 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr2_+_19522082 | 0.30 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr2_+_20793982 | 0.30 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr9_-_9415000 | 0.30 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr22_+_19552987 | 0.30 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr8_+_25034544 | 0.30 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr7_-_67248829 | 0.30 |
ENSDART00000192442
|
znf143a
|
zinc finger protein 143a |
| chr25_-_10791437 | 0.30 |
ENSDART00000127054
|
BX572619.1
|
|
| chr11_+_33818179 | 0.29 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr21_-_5066462 | 0.28 |
ENSDART00000067733
|
zgc:77838
|
zgc:77838 |
| chr7_+_9873750 | 0.28 |
ENSDART00000173348
ENSDART00000172813 |
lins1
|
lines homolog 1 |
| chr17_+_15433671 | 0.28 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr1_-_5455498 | 0.28 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr24_+_9412450 | 0.28 |
ENSDART00000132724
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr20_-_9095105 | 0.27 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr9_+_8396755 | 0.27 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr13_+_27232848 | 0.27 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr9_-_25181137 | 0.27 |
ENSDART00000192624
|
esd
|
esterase D/formylglutathione hydrolase |
| chr9_-_12424791 | 0.26 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr9_+_50001746 | 0.26 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr12_+_47698356 | 0.25 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr17_-_31639845 | 0.25 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr21_+_45502621 | 0.24 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr3_+_28860283 | 0.24 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr22_-_19552796 | 0.24 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr12_+_33817327 | 0.23 |
ENSDART00000111486
|
mrpl43
|
mitochondrial ribosomal protein L43 |
| chr15_-_16177603 | 0.23 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr2_+_19633493 | 0.23 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr15_-_21877726 | 0.23 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr22_-_20166660 | 0.23 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr22_+_2937485 | 0.22 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr7_+_20471315 | 0.22 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr20_+_46492175 | 0.22 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr8_+_21353878 | 0.22 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr15_-_22074315 | 0.22 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr5_-_67750907 | 0.22 |
ENSDART00000172097
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr9_+_37329036 | 0.22 |
ENSDART00000131756
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr19_+_7152966 | 0.22 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr25_+_35913614 | 0.22 |
ENSDART00000022437
|
gpia
|
glucose-6-phosphate isomerase a |
| chr5_+_36415978 | 0.22 |
ENSDART00000084464
|
fam155b
|
family with sequence similarity 155, member B |
| chr11_-_45138857 | 0.21 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr20_-_20930926 | 0.21 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr5_+_42400777 | 0.21 |
ENSDART00000183114
|
BX548073.8
|
|
| chr2_-_19520324 | 0.19 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr25_-_3759322 | 0.19 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr8_-_37249813 | 0.19 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr11_-_6048490 | 0.19 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr19_+_41464870 | 0.19 |
ENSDART00000102778
|
dlx6a
|
distal-less homeobox 6a |
| chr11_-_40728380 | 0.19 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr5_-_42661012 | 0.19 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr2_+_37227011 | 0.18 |
ENSDART00000126587
ENSDART00000084958 |
samd7
|
sterile alpha motif domain containing 7 |
| chr11_+_35050253 | 0.18 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr2_+_36721357 | 0.18 |
ENSDART00000019063
|
pdcd10b
|
programmed cell death 10b |
| chr20_-_45060241 | 0.18 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr6_+_52350443 | 0.18 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr9_-_41077934 | 0.18 |
ENSDART00000100342
|
ankar
|
ankyrin and armadillo repeat containing |
| chr19_-_42391383 | 0.18 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr2_+_14992879 | 0.18 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr17_-_42492668 | 0.17 |
ENSDART00000183946
|
CR925755.2
|
|
| chr25_+_3035384 | 0.17 |
ENSDART00000184219
ENSDART00000149360 |
mpi
|
mannose phosphate isomerase |
| chr10_+_43039947 | 0.17 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr2_+_19578446 | 0.17 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr6_-_8311044 | 0.17 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr11_-_37997419 | 0.17 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr7_-_30174882 | 0.17 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr9_-_31278048 | 0.17 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr14_-_24761132 | 0.17 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr10_-_11261386 | 0.16 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr8_-_37249991 | 0.16 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr5_-_41494831 | 0.16 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr10_+_11767791 | 0.16 |
ENSDART00000092047
|
ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr17_+_24722646 | 0.16 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr6_-_51386656 | 0.16 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr22_-_382955 | 0.16 |
ENSDART00000082406
|
ora3
|
olfactory receptor class A related 3 |
| chr18_-_14677936 | 0.16 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr20_-_20931197 | 0.16 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr17_-_29119362 | 0.16 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr25_-_27621268 | 0.15 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr6_+_3280939 | 0.15 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr3_+_13929860 | 0.15 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr13_+_22295905 | 0.15 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr5_-_12219572 | 0.15 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr22_-_15578402 | 0.15 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr11_+_17984167 | 0.15 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr1_-_31171242 | 0.15 |
ENSDART00000190294
|
kcnq5b
|
potassium voltage-gated channel, KQT-like subfamily, member 5b |
| chr5_+_66433287 | 0.15 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr21_-_40676224 | 0.14 |
ENSDART00000162623
|
arxb
|
aristaless related homeobox b |
| chr7_-_6445129 | 0.14 |
ENSDART00000172825
|
FP325123.2
|
Histone H3.2 |
| chr1_-_10821584 | 0.14 |
ENSDART00000167452
ENSDART00000162137 |
crfb15
|
cytokine receptor family member B15 |
| chr20_+_18551657 | 0.14 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr9_+_34641237 | 0.14 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr9_-_51436377 | 0.14 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr10_-_32494304 | 0.14 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr16_-_55028740 | 0.14 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr4_-_16334362 | 0.14 |
ENSDART00000101461
|
epyc
|
epiphycan |
| chr12_-_6880694 | 0.14 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr20_-_14925281 | 0.14 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr6_+_41191482 | 0.14 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr9_+_38163876 | 0.14 |
ENSDART00000137955
|
clasp1a
|
cytoplasmic linker associated protein 1a |
| chr16_-_42894628 | 0.14 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr21_+_42226113 | 0.14 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr17_+_43032529 | 0.14 |
ENSDART00000055611
ENSDART00000154863 |
isca2
|
iron-sulfur cluster assembly 2 |
| chr17_-_36896560 | 0.13 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr6_-_20952187 | 0.13 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr24_+_16985181 | 0.13 |
ENSDART00000135580
|
eif2s3
|
eukaryotic translation initiation factor 2, subunit 3 gamma |
| chr23_-_31974060 | 0.13 |
ENSDART00000168087
|
BX927210.1
|
|
| chr24_-_17067284 | 0.13 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr17_-_31579715 | 0.13 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr3_+_46635527 | 0.13 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr16_+_23303859 | 0.13 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr10_-_34871737 | 0.13 |
ENSDART00000138755
|
dclk1a
|
doublecortin-like kinase 1a |
| chr23_+_24954951 | 0.13 |
ENSDART00000136941
|
nol9
|
nucleolar protein 9 |
| chr13_-_25720876 | 0.13 |
ENSDART00000142404
ENSDART00000146487 |
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr24_+_21540842 | 0.12 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr17_+_10593398 | 0.12 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
| chr18_-_20869175 | 0.12 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr17_+_6625717 | 0.12 |
ENSDART00000190753
ENSDART00000181298 |
BX005321.1
|
|
| chr1_-_44434707 | 0.12 |
ENSDART00000110148
|
cryba1l2
|
crystallin, beta A1, like 2 |
| chr8_+_30112655 | 0.12 |
ENSDART00000099027
|
fancc
|
Fanconi anemia, complementation group C |
| chr24_+_35787629 | 0.12 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr13_-_12602920 | 0.12 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr9_-_19161982 | 0.12 |
ENSDART00000081878
|
pou1f1
|
POU class 1 homeobox 1 |
| chr4_+_16885854 | 0.12 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr20_+_25586099 | 0.12 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr16_+_5774977 | 0.12 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr22_+_7439186 | 0.12 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr6_-_55254786 | 0.12 |
ENSDART00000113805
|
nfatc2b
|
nuclear factor of activated T cells 2b |
| chr10_-_32494499 | 0.12 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr3_-_41791178 | 0.12 |
ENSDART00000049687
|
grifin
|
galectin-related inter-fiber protein |
| chr2_-_30668580 | 0.12 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr13_+_25449681 | 0.11 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr15_-_14552101 | 0.11 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr6_+_13117598 | 0.11 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr25_-_13490744 | 0.11 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr21_-_22827548 | 0.11 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr19_+_20274491 | 0.11 |
ENSDART00000090860
ENSDART00000151693 |
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr20_+_539852 | 0.11 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr20_-_14924858 | 0.11 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of lmx1ba+lmx1bb_lmx1al
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 0.6 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 0.7 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.1 | 0.8 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.1 | 0.5 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 1.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.1 | 0.2 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.0 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.1 | 0.2 | GO:0044038 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.3 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.8 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.1 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.1 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.6 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.6 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.0 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.5 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.2 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.8 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.8 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.2 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.5 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.2 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.3 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.2 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.1 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |