Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
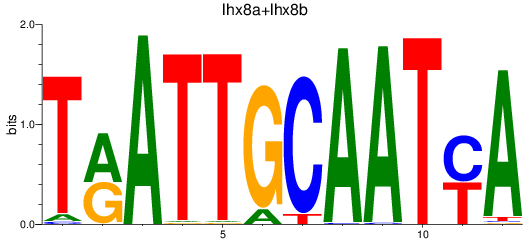
Results for lhx8a+lhx8b
Z-value: 0.66
Transcription factors associated with lhx8a+lhx8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx8a
|
ENSDARG00000002330 | LIM homeobox 8a |
|
lhx8b
|
ENSDARG00000042145 | LIM homeobox 8b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx8a | dr11_v1_chr2_+_11205795_11205795 | -0.64 | 4.1e-03 | Click! |
| lhx8b | dr11_v1_chr8_-_17926620_17926620 | 0.23 | 3.6e-01 | Click! |
Activity profile of lhx8a+lhx8b motif
Sorted Z-values of lhx8a+lhx8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_32384683 | 0.95 |
ENSDART00000182942
ENSDART00000141757 |
ubtfl
|
upstream binding transcription factor, like |
| chr11_+_13424116 | 0.86 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr11_+_13423776 | 0.66 |
ENSDART00000102553
|
homer3b
|
homer scaffolding protein 3b |
| chr8_+_29635968 | 0.61 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr11_-_43200994 | 0.59 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr17_+_10570906 | 0.57 |
ENSDART00000182371
ENSDART00000176097 ENSDART00000110593 ENSDART00000169356 |
mgaa
|
MGA, MAX dimerization protein a |
| chr21_-_36571804 | 0.57 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr10_+_20364009 | 0.57 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr8_+_29636431 | 0.56 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr4_+_14727018 | 0.56 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr18_-_158541 | 0.55 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr2_+_49644803 | 0.52 |
ENSDART00000160342
|
si:ch211-209f23.7
|
si:ch211-209f23.7 |
| chr4_+_14727212 | 0.51 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr7_+_13609457 | 0.50 |
ENSDART00000172857
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr20_-_7080427 | 0.49 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr7_+_41887429 | 0.49 |
ENSDART00000115090
|
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr15_-_34866219 | 0.49 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr16_-_36979592 | 0.48 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr15_+_11427620 | 0.47 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr8_+_10823069 | 0.46 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr16_-_21903083 | 0.45 |
ENSDART00000165849
|
setdb1b
|
SET domain, bifurcated 1b |
| chr15_-_34865952 | 0.44 |
ENSDART00000186868
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr21_-_13662237 | 0.44 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr19_+_46222428 | 0.44 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr1_+_45671687 | 0.44 |
ENSDART00000146101
|
mcoln1a
|
mucolipin 1a |
| chr20_+_6535438 | 0.44 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr7_-_39378903 | 0.42 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr23_-_29668286 | 0.42 |
ENSDART00000129248
|
clstn1
|
calsyntenin 1 |
| chr25_+_19238175 | 0.42 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr24_+_39027481 | 0.41 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr9_-_12401871 | 0.40 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr5_+_44944778 | 0.40 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr5_-_15494164 | 0.40 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr13_+_29925397 | 0.39 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr4_-_5775507 | 0.38 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr16_-_35329803 | 0.38 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr17_-_29271359 | 0.37 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr25_-_36263115 | 0.36 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr21_-_34926619 | 0.36 |
ENSDART00000065337
ENSDART00000192740 |
kif20a
|
kinesin family member 20A |
| chr3_-_33935200 | 0.35 |
ENSDART00000151211
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr14_-_23801389 | 0.35 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_+_28518349 | 0.34 |
ENSDART00000159961
|
stag2b
|
stromal antigen 2b |
| chr17_+_15559046 | 0.34 |
ENSDART00000187126
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr5_+_36654817 | 0.34 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr7_-_32629458 | 0.34 |
ENSDART00000001376
|
arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr9_+_42606681 | 0.34 |
ENSDART00000191186
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr3_-_30488063 | 0.34 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr4_+_9536860 | 0.33 |
ENSDART00000130083
|
lsm8
|
LSM8 homolog, U6 small nuclear RNA associated |
| chr25_+_21098675 | 0.33 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr17_-_33412868 | 0.33 |
ENSDART00000187521
|
BX323819.1
|
|
| chr8_-_16725959 | 0.32 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr14_-_33945692 | 0.32 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr6_+_2195625 | 0.32 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr24_+_12945803 | 0.32 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr14_+_8638353 | 0.32 |
ENSDART00000163240
|
si:dkeyp-115e12.6
|
si:dkeyp-115e12.6 |
| chr8_-_16725573 | 0.32 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr24_+_792429 | 0.31 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr21_+_27302752 | 0.31 |
ENSDART00000012855
|
sart1
|
SART1, U4/U6.U5 tri-snRNP-associated protein 1 |
| chr13_-_8892514 | 0.30 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr1_-_55873178 | 0.30 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr3_-_33934788 | 0.29 |
ENSDART00000151411
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr9_+_42607138 | 0.29 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr5_+_13326765 | 0.29 |
ENSDART00000090851
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr9_+_25775816 | 0.28 |
ENSDART00000127834
ENSDART00000189994 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr24_-_32025637 | 0.28 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr13_+_29292011 | 0.28 |
ENSDART00000115023
|
parga
|
poly (ADP-ribose) glycohydrolase a |
| chr5_+_57320113 | 0.28 |
ENSDART00000036331
|
atp6v1g1
|
ATPase H+ transporting V1 subunit G1 |
| chr4_+_15605844 | 0.27 |
ENSDART00000101619
ENSDART00000021384 |
exoc4
|
exocyst complex component 4 |
| chr17_+_24590177 | 0.27 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr19_-_31042570 | 0.27 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr17_+_41081512 | 0.27 |
ENSDART00000056742
|
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr1_-_7673376 | 0.27 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr18_+_26899316 | 0.27 |
ENSDART00000050230
|
tspan3a
|
tetraspanin 3a |
| chr21_-_44771213 | 0.26 |
ENSDART00000188534
|
kif4
|
kinesin family member 4 |
| chr22_-_910926 | 0.26 |
ENSDART00000180075
|
FP016205.1
|
|
| chr22_-_15562933 | 0.25 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr24_+_33589667 | 0.25 |
ENSDART00000152097
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr20_-_33507458 | 0.25 |
ENSDART00000140287
|
paplnb
|
papilin b, proteoglycan-like sulfated glycoprotein |
| chr10_+_17747880 | 0.25 |
ENSDART00000135044
|
pigo
|
phosphatidylinositol glycan anchor biosynthesis, class O |
| chr16_-_35427060 | 0.25 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr2_+_50062165 | 0.24 |
ENSDART00000097939
ENSDART00000138214 |
zcchc2
|
zinc finger, CCHC domain containing 2 |
| chr1_+_10019653 | 0.24 |
ENSDART00000190227
|
trim2b
|
tripartite motif containing 2b |
| chr19_+_366034 | 0.24 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr13_-_24260609 | 0.24 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr6_-_16456093 | 0.24 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr17_+_21486047 | 0.24 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr15_-_25613114 | 0.23 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr13_+_8892784 | 0.23 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr24_+_39137001 | 0.23 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr2_-_22659450 | 0.23 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr15_-_45356635 | 0.23 |
ENSDART00000192000
|
CABZ01068246.1
|
|
| chr11_+_36355348 | 0.23 |
ENSDART00000145427
|
sypl2a
|
synaptophysin-like 2a |
| chr7_-_51727760 | 0.23 |
ENSDART00000174180
|
hdac8
|
histone deacetylase 8 |
| chr15_-_5901514 | 0.22 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr6_-_12456077 | 0.22 |
ENSDART00000190107
|
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr7_-_30280934 | 0.22 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr25_+_15938880 | 0.21 |
ENSDART00000089035
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr9_-_16218097 | 0.21 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr22_+_1313046 | 0.21 |
ENSDART00000170421
|
si:ch73-138e16.6
|
si:ch73-138e16.6 |
| chr3_-_45281350 | 0.20 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr21_-_25601648 | 0.20 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr21_-_8153165 | 0.20 |
ENSDART00000182580
|
CABZ01074363.1
|
|
| chr7_-_64770456 | 0.20 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr16_+_25535993 | 0.19 |
ENSDART00000077436
|
mylipb
|
myosin regulatory light chain interacting protein b |
| chr21_+_21201346 | 0.19 |
ENSDART00000142961
|
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr5_-_8614217 | 0.19 |
ENSDART00000164844
|
rictora
|
RPTOR independent companion of MTOR, complex 2 a |
| chr13_+_43650632 | 0.19 |
ENSDART00000141024
|
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr6_+_40661703 | 0.18 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr7_+_72469724 | 0.18 |
ENSDART00000187629
|
HECTD4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr11_+_44236183 | 0.18 |
ENSDART00000193470
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr1_+_7414318 | 0.18 |
ENSDART00000127426
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr8_+_7737062 | 0.18 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr12_-_24812403 | 0.17 |
ENSDART00000185517
|
foxn2b
|
forkhead box N2b |
| chr3_+_50201240 | 0.17 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr8_+_3820134 | 0.16 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr7_-_2285499 | 0.16 |
ENSDART00000182211
|
si:dkey-187j14.6
|
si:dkey-187j14.6 |
| chr9_-_25055722 | 0.16 |
ENSDART00000137131
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr17_-_30652738 | 0.16 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr2_-_14387335 | 0.16 |
ENSDART00000189332
ENSDART00000164786 ENSDART00000188572 |
sgip1b
|
SH3-domain GRB2-like (endophilin) interacting protein 1b |
| chr9_-_41040492 | 0.16 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr16_+_29514473 | 0.16 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr16_+_4838808 | 0.15 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr8_-_23612462 | 0.15 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr17_-_22552678 | 0.15 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr19_-_43004356 | 0.15 |
ENSDART00000098101
|
zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr20_+_492529 | 0.15 |
ENSDART00000141709
|
FO904980.1
|
|
| chr21_+_43400351 | 0.15 |
ENSDART00000141098
|
frmd7
|
FERM domain containing 7 |
| chr11_-_12471837 | 0.15 |
ENSDART00000113175
ENSDART00000186965 |
si:dkey-27d5.6
|
si:dkey-27d5.6 |
| chr20_+_34151670 | 0.14 |
ENSDART00000152870
ENSDART00000010329 ENSDART00000145852 |
arpc5b
|
actin related protein 2/3 complex, subunit 5B |
| chr17_+_7513673 | 0.14 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr21_-_17016788 | 0.14 |
ENSDART00000186703
|
AL844518.1
|
|
| chr18_+_30370559 | 0.14 |
ENSDART00000165450
|
gse1
|
Gse1 coiled-coil protein |
| chr15_-_45246911 | 0.14 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr25_+_28893615 | 0.14 |
ENSDART00000156994
ENSDART00000075151 |
amn1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr10_-_44482911 | 0.14 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr23_-_19051710 | 0.14 |
ENSDART00000111852
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr8_+_8196087 | 0.14 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr5_+_27137473 | 0.14 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr6_+_19948043 | 0.14 |
ENSDART00000182636
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr13_+_21677767 | 0.13 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr15_-_41708657 | 0.13 |
ENSDART00000154384
|
si:ch211-276c2.4
|
si:ch211-276c2.4 |
| chr11_+_11430725 | 0.13 |
ENSDART00000188040
|
si:dkey-23f9.11
|
si:dkey-23f9.11 |
| chr4_-_25812329 | 0.13 |
ENSDART00000146658
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr4_+_612363 | 0.13 |
ENSDART00000049154
|
pthlha
|
parathyroid hormone-like hormone a |
| chr2_+_42005217 | 0.13 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr25_+_7591293 | 0.13 |
ENSDART00000130416
|
FO704779.1
|
|
| chr21_-_17016105 | 0.12 |
ENSDART00000189120
|
AL844518.1
|
|
| chr21_-_11970199 | 0.12 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr19_-_20162980 | 0.12 |
ENSDART00000184605
|
fam221a
|
family with sequence similarity 221, member A |
| chr7_-_1101694 | 0.11 |
ENSDART00000183017
ENSDART00000182681 |
dctn1a
|
dynactin 1a |
| chr4_-_25515154 | 0.11 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr5_-_20205075 | 0.11 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr23_-_19051869 | 0.11 |
ENSDART00000140866
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr10_-_36721726 | 0.11 |
ENSDART00000122132
|
pimr137
|
Pim proto-oncogene, serine/threonine kinase, related 137 |
| chr3_-_52674089 | 0.11 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr22_-_817479 | 0.11 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr24_+_26887487 | 0.11 |
ENSDART00000189425
|
CR376848.1
|
|
| chr7_+_16509201 | 0.10 |
ENSDART00000173777
|
zdhhc13
|
zinc finger, DHHC-type containing 13 |
| chr5_+_51026563 | 0.10 |
ENSDART00000050988
|
gcnt4a
|
glucosaminyl (N-acetyl) transferase 4, core 2, a |
| chr23_+_32101361 | 0.10 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr8_+_20438884 | 0.10 |
ENSDART00000016422
ENSDART00000133794 |
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr10_+_14210667 | 0.10 |
ENSDART00000138854
|
si:dkey-286h21.1
|
si:dkey-286h21.1 |
| chr23_+_45282858 | 0.10 |
ENSDART00000162353
|
CABZ01073265.1
|
|
| chr16_-_24819860 | 0.10 |
ENSDART00000183874
|
kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr15_+_36966369 | 0.09 |
ENSDART00000163622
|
kirrel3l
|
kirre like nephrin family adhesion molecule 3, like |
| chr10_-_43568239 | 0.09 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr6_+_11249706 | 0.09 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr9_+_25776971 | 0.09 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr9_-_16853462 | 0.09 |
ENSDART00000160273
|
CT573248.2
|
|
| chr11_-_12350237 | 0.09 |
ENSDART00000191094
|
si:dkey-27d5.14
|
si:dkey-27d5.14 |
| chr9_-_2892250 | 0.09 |
ENSDART00000140695
|
cdca7a
|
cell division cycle associated 7a |
| chr23_+_21278948 | 0.08 |
ENSDART00000156701
ENSDART00000033970 |
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr18_-_48558420 | 0.08 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr6_+_12865137 | 0.08 |
ENSDART00000090065
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr20_+_18551657 | 0.08 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr20_+_42563618 | 0.08 |
ENSDART00000153441
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr15_+_5132439 | 0.08 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr23_-_43486714 | 0.08 |
ENSDART00000169726
|
e2f1
|
E2F transcription factor 1 |
| chr4_+_3455146 | 0.08 |
ENSDART00000180124
|
znf800b
|
zinc finger protein 800b |
| chr24_+_9003998 | 0.07 |
ENSDART00000179656
ENSDART00000191314 |
CR318624.2
|
|
| chr15_+_32268790 | 0.07 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr9_-_21918963 | 0.07 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr20_+_23173710 | 0.07 |
ENSDART00000074172
|
sgcb
|
sarcoglycan, beta (dystrophin-associated glycoprotein) |
| chr23_+_36095260 | 0.07 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr4_-_22311610 | 0.07 |
ENSDART00000137814
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr9_-_2892045 | 0.07 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr1_-_2457546 | 0.07 |
ENSDART00000103795
|
ggact.1
|
gamma-glutamylamine cyclotransferase, tandem duplicate 1 |
| chr11_+_24620742 | 0.06 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr15_-_1038193 | 0.06 |
ENSDART00000159462
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr16_+_20926673 | 0.06 |
ENSDART00000009827
|
hoxa2b
|
homeobox A2b |
| chr14_-_25577094 | 0.06 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr13_+_10970614 | 0.06 |
ENSDART00000058147
|
dync2li1
|
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr6_+_20647155 | 0.06 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr25_-_28768489 | 0.06 |
ENSDART00000088315
|
washc4
|
WASH complex subunit 4 |
| chr22_+_10010292 | 0.06 |
ENSDART00000180096
|
BX324216.4
|
|
| chr10_+_24631976 | 0.06 |
ENSDART00000181928
|
slc46a3
|
solute carrier family 46, member 3 |
| chr11_+_25276748 | 0.05 |
ENSDART00000126211
|
cyldb
|
cylindromatosis (turban tumor syndrome), b |
| chr7_-_31441420 | 0.05 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr7_-_22941472 | 0.05 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr7_+_53152108 | 0.05 |
ENSDART00000171350
|
cdh29
|
cadherin 29 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx8a+lhx8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 0.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.4 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 1.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 1.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.1 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.9 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.7 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.4 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.4 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.4 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.1 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) ectodermal cell fate specification(GO:0001715) ectodermal cell differentiation(GO:0010668) regulation of ectodermal cell fate specification(GO:0042665) regulation of ectoderm development(GO:2000383) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.3 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.4 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.3 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.2 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 0.6 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.3 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.2 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.6 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0097526 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.4 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.6 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 0.4 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 1.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.1 | 0.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.4 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.0 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.9 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 1.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |