Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
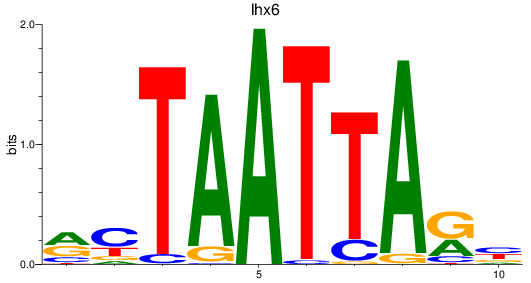
Results for lhx6
Z-value: 0.64
Transcription factors associated with lhx6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6
|
ENSDARG00000006896 | LIM homeobox 6 |
|
lhx6
|
ENSDARG00000112520 | LIM homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx6 | dr11_v1_chr10_+_9372702_9372702 | -0.93 | 3.1e-08 | Click! |
Activity profile of lhx6 motif
Sorted Z-values of lhx6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_21362071 | 3.35 |
ENSDART00000125167
|
avd
|
avidin |
| chr8_-_23780334 | 3.26 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr10_-_21362320 | 3.13 |
ENSDART00000189789
|
avd
|
avidin |
| chr10_-_34002185 | 2.47 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr21_+_25777425 | 2.28 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr11_-_44801968 | 2.25 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr12_-_14143344 | 2.19 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr12_-_33357655 | 2.14 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_-_35633827 | 1.96 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr2_-_15324837 | 1.91 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr23_+_2728095 | 1.74 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr6_+_28208973 | 1.72 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr19_+_31585917 | 1.68 |
ENSDART00000132182
|
gmnn
|
geminin, DNA replication inhibitor |
| chr24_-_9979342 | 1.62 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr5_+_37903790 | 1.60 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr2_+_6253246 | 1.58 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr25_+_5972690 | 1.53 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr20_-_6532462 | 1.49 |
ENSDART00000054653
|
mcm3l
|
MCM3 minichromosome maintenance deficient 3 (S. cerevisiae), like |
| chr15_+_46313082 | 1.38 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr20_-_14114078 | 1.37 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr20_+_14114258 | 1.35 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr1_-_18811517 | 1.34 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr6_+_21001264 | 1.33 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr12_-_48188928 | 1.30 |
ENSDART00000184384
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr20_-_23426339 | 1.26 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr21_-_32060993 | 1.17 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr1_-_55248496 | 1.06 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr17_-_40956035 | 1.04 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr10_-_34916208 | 1.00 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr13_+_38814521 | 0.96 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr14_+_8940326 | 0.95 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr11_+_37638873 | 0.93 |
ENSDART00000186384
ENSDART00000184291 ENSDART00000131782 ENSDART00000140502 |
sh2d5
|
SH2 domain containing 5 |
| chr14_+_15155684 | 0.92 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr7_-_59159253 | 0.92 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr2_-_26596794 | 0.89 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_+_11261576 | 0.88 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr23_-_28120058 | 0.88 |
ENSDART00000087815
|
b4galnt1a
|
beta-1,4-N-acetyl-galactosaminyl transferase 1a |
| chr17_+_16046314 | 0.88 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr7_+_51795667 | 0.87 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr14_+_34492288 | 0.86 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr8_-_25034411 | 0.78 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr19_+_10661520 | 0.77 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr20_+_29209767 | 0.76 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr16_+_47207691 | 0.76 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr5_+_25733774 | 0.74 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr23_-_10786400 | 0.74 |
ENSDART00000055038
|
rybpa
|
RING1 and YY1 binding protein a |
| chr25_-_6049339 | 0.74 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr10_+_33393829 | 0.73 |
ENSDART00000163458
ENSDART00000115379 |
zgc:153345
|
zgc:153345 |
| chr4_+_4849789 | 0.72 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr8_-_25033681 | 0.72 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr6_+_50393047 | 0.72 |
ENSDART00000055502
ENSDART00000055511 |
ergic3
|
ERGIC and golgi 3 |
| chr1_-_513762 | 0.71 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr15_+_28410664 | 0.71 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr20_+_29209926 | 0.70 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr9_-_11676491 | 0.70 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr5_-_19006290 | 0.70 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr7_-_26532089 | 0.69 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr14_+_35428152 | 0.69 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr11_+_18873619 | 0.68 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr1_-_23308225 | 0.67 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr10_-_35257458 | 0.66 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr8_-_15129573 | 0.66 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr22_-_21897203 | 0.66 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr23_-_33709964 | 0.66 |
ENSDART00000143333
ENSDART00000130338 |
pou6f1
|
POU class 6 homeobox 1 |
| chr5_+_29851433 | 0.65 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr13_-_35808904 | 0.64 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr15_+_17345609 | 0.62 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr9_-_746317 | 0.62 |
ENSDART00000129632
ENSDART00000130720 |
usp37
|
ubiquitin specific peptidase 37 |
| chr24_+_29912509 | 0.62 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr23_-_36446307 | 0.62 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr20_+_29209615 | 0.61 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr5_+_60590796 | 0.61 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr1_+_13930625 | 0.61 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr6_-_12172424 | 0.60 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr17_+_37227936 | 0.59 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr7_-_49594995 | 0.58 |
ENSDART00000174161
ENSDART00000109147 |
brsk2b
|
BR serine/threonine kinase 2b |
| chr12_-_35393211 | 0.57 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr1_+_35985813 | 0.56 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr24_+_19415124 | 0.55 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr15_-_18115540 | 0.54 |
ENSDART00000131639
ENSDART00000047902 |
arcn1b
|
archain 1b |
| chr12_+_22580579 | 0.53 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr8_-_49728590 | 0.52 |
ENSDART00000135714
ENSDART00000138810 ENSDART00000098319 |
gkap1
|
G kinase anchoring protein 1 |
| chr4_-_2637689 | 0.52 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr5_-_25733745 | 0.52 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr13_+_38817871 | 0.52 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr5_+_36768674 | 0.51 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr25_-_10791437 | 0.51 |
ENSDART00000127054
|
BX572619.1
|
|
| chr4_+_2637947 | 0.50 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr2_+_26237322 | 0.50 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr1_-_40102836 | 0.50 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr1_+_513986 | 0.49 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr14_-_16082806 | 0.49 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr21_-_18275226 | 0.48 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr22_-_14247276 | 0.48 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr23_-_2901167 | 0.47 |
ENSDART00000165955
ENSDART00000190616 |
zhx3
|
zinc fingers and homeoboxes 3 |
| chr12_-_18577983 | 0.47 |
ENSDART00000193262
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr25_+_35891342 | 0.46 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr24_-_25004553 | 0.46 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr20_-_9436521 | 0.45 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr21_-_39177564 | 0.45 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr23_-_29394505 | 0.44 |
ENSDART00000017728
|
pgd
|
phosphogluconate dehydrogenase |
| chr3_-_26787430 | 0.43 |
ENSDART00000087047
|
rab40c
|
RAB40c, member RAS oncogene family |
| chr20_-_37813863 | 0.43 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr17_+_49281597 | 0.42 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr8_-_23776399 | 0.42 |
ENSDART00000114800
|
INAVA
|
si:ch211-163l21.4 |
| chr2_+_40294313 | 0.41 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr5_-_67241633 | 0.41 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr19_-_12648122 | 0.40 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr16_-_41714988 | 0.40 |
ENSDART00000138798
|
cep85
|
centrosomal protein 85 |
| chr15_-_9272328 | 0.40 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr9_+_18829360 | 0.39 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr15_+_21262917 | 0.39 |
ENSDART00000101000
|
gkup
|
glucuronokinase with putative uridyl pyrophosphorylase |
| chr4_-_77557279 | 0.39 |
ENSDART00000180113
|
AL935186.10
|
|
| chr23_+_27782071 | 0.38 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr6_-_55585423 | 0.38 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr17_+_8799661 | 0.38 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr4_+_25917915 | 0.37 |
ENSDART00000138603
|
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr8_-_21142550 | 0.37 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr10_-_32494499 | 0.35 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr4_-_73756673 | 0.35 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr9_-_50001606 | 0.35 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr2_-_38363017 | 0.34 |
ENSDART00000088026
|
prmt5
|
protein arginine methyltransferase 5 |
| chr17_-_43594864 | 0.34 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr20_-_48604199 | 0.34 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr25_-_29087925 | 0.34 |
ENSDART00000171758
|
rpp25a
|
ribonuclease P and MRP subunit p25, a |
| chr9_-_3934963 | 0.33 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr20_+_28803977 | 0.33 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr3_+_27798094 | 0.32 |
ENSDART00000075100
ENSDART00000151437 |
carhsp1
|
calcium regulated heat stable protein 1 |
| chr6_-_40922971 | 0.32 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr3_+_53116172 | 0.32 |
ENSDART00000115117
|
brd4
|
bromodomain containing 4 |
| chr6_-_3982783 | 0.32 |
ENSDART00000171944
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr13_-_31017960 | 0.31 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr17_+_8799451 | 0.31 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr21_+_20386865 | 0.31 |
ENSDART00000144366
|
si:dkey-30k6.5
|
si:dkey-30k6.5 |
| chr6_-_40713183 | 0.31 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr10_-_42519773 | 0.31 |
ENSDART00000039187
|
march5l
|
membrane-associated ring finger (C3HC4) 5, like |
| chr7_+_36898850 | 0.31 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr7_+_17908235 | 0.31 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr13_+_2442841 | 0.30 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr13_+_40501455 | 0.30 |
ENSDART00000114985
|
hpse2
|
heparanase 2 |
| chr4_-_4834617 | 0.30 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr24_+_16547035 | 0.30 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr19_-_12648408 | 0.30 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr9_-_28274932 | 0.29 |
ENSDART00000137582
ENSDART00000146932 |
creb1b
|
cAMP responsive element binding protein 1b |
| chr5_+_19933356 | 0.29 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr18_-_15551360 | 0.29 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr14_+_29941445 | 0.29 |
ENSDART00000181761
|
fam149a
|
family with sequence similarity 149 member A |
| chr3_+_45364849 | 0.29 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr10_-_32494304 | 0.29 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr5_-_68093169 | 0.27 |
ENSDART00000051849
|
slc25a11
|
solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 |
| chr3_-_43356082 | 0.27 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr21_+_18274825 | 0.27 |
ENSDART00000144322
ENSDART00000147768 |
wdr5
|
WD repeat domain 5 |
| chr18_+_17725410 | 0.26 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr20_-_34750363 | 0.26 |
ENSDART00000152845
|
znf395b
|
zinc finger protein 395b |
| chr6_-_48418021 | 0.26 |
ENSDART00000090528
|
rhoca
|
ras homolog family member Ca |
| chr1_-_21563040 | 0.25 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr13_+_15838151 | 0.25 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr24_+_39518774 | 0.25 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr3_-_61162750 | 0.25 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr9_+_50001746 | 0.25 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr7_-_30174882 | 0.24 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr19_+_47502581 | 0.24 |
ENSDART00000171526
|
CU695215.2
|
|
| chr8_+_25034544 | 0.24 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr1_-_15797663 | 0.24 |
ENSDART00000177122
|
SGCZ
|
sarcoglycan zeta |
| chr12_+_48803098 | 0.23 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr4_-_4834347 | 0.23 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr21_+_42226113 | 0.23 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr11_+_31864921 | 0.22 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr16_+_33144306 | 0.22 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr21_+_34814444 | 0.22 |
ENSDART00000161816
|
wdr55
|
WD repeat domain 55 |
| chr9_-_43538328 | 0.22 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr9_+_17983463 | 0.22 |
ENSDART00000182150
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr2_+_37140448 | 0.22 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr5_+_61301525 | 0.21 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr23_-_46020226 | 0.21 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr11_+_5588122 | 0.21 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr17_-_22552678 | 0.20 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr4_+_3980247 | 0.20 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr5_-_69004007 | 0.20 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr10_+_13000669 | 0.20 |
ENSDART00000158919
ENSDART00000172625 |
lpar1
|
lysophosphatidic acid receptor 1 |
| chr24_-_39518599 | 0.19 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr5_-_15851953 | 0.19 |
ENSDART00000173101
|
si:dkey-1k23.3
|
si:dkey-1k23.3 |
| chr21_+_26748141 | 0.19 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr20_+_41021054 | 0.18 |
ENSDART00000146052
|
man1a1
|
mannosidase, alpha, class 1A, member 1 |
| chr20_-_34750045 | 0.18 |
ENSDART00000186130
|
znf395b
|
zinc finger protein 395b |
| chr16_-_26855936 | 0.18 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr7_-_12464412 | 0.18 |
ENSDART00000178723
|
adamtsl3
|
ADAMTS-like 3 |
| chr10_-_11385155 | 0.18 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr8_-_49766205 | 0.17 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr18_+_2554666 | 0.17 |
ENSDART00000167218
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr13_+_38521152 | 0.17 |
ENSDART00000145292
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr22_-_20166660 | 0.17 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr7_-_7692723 | 0.17 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr17_-_37395460 | 0.17 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr23_-_18130264 | 0.16 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr19_-_42588510 | 0.16 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr9_-_51436377 | 0.16 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr9_+_19529951 | 0.16 |
ENSDART00000125416
|
pknox1.1
|
pbx/knotted 1 homeobox 1.1 |
| chr17_+_20589553 | 0.15 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr13_-_38088627 | 0.15 |
ENSDART00000175268
|
CT027676.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 0.9 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.2 | 1.7 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 1.5 | GO:1902315 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.2 | 0.9 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.7 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.5 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 2.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 0.4 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.8 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.3 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.1 | 1.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.1 | 0.6 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.6 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.8 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.2 | GO:0070257 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.3 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.4 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.7 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.4 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.4 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 1.0 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.6 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0060465 | pharynx development(GO:0060465) |
| 0.0 | 0.3 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.9 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 2.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.2 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 1.0 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 1.5 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.6 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.6 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 1.3 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.1 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.0 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.3 | 0.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 0.9 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 1.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.7 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 2.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.3 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 2.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.4 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 0.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.6 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 2.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.7 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 1.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.8 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 1.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.3 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 0.4 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 2.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.3 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.8 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.0 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 1.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |