Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
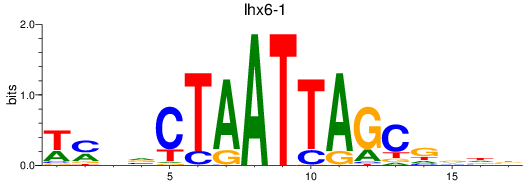
Results for lhx6-1
Z-value: 0.59
Transcription factors associated with lhx6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx6-1
|
ENSDARG00000052165 | si_ch211-236k19.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-236k19.2 | dr11_v1_chr5_-_64900552_64900552 | -0.49 | 3.9e-02 | Click! |
Activity profile of lhx6-1 motif
Sorted Z-values of lhx6-1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_28208973 | 2.17 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr12_-_33354409 | 1.75 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_38284648 | 1.72 |
ENSDART00000148281
ENSDART00000132621 |
si:ch211-14a17.7
|
si:ch211-14a17.7 |
| chr9_+_44994214 | 1.49 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr12_-_33357655 | 1.48 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr6_-_8360918 | 1.39 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr24_-_9979342 | 1.32 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr2_+_6255434 | 1.25 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr7_+_51795667 | 1.23 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr8_-_23780334 | 1.18 |
ENSDART00000145179
ENSDART00000145894 |
zgc:195245
|
zgc:195245 |
| chr5_+_37903790 | 1.18 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr24_-_24724233 | 1.17 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr11_-_16021424 | 1.11 |
ENSDART00000193291
ENSDART00000170731 ENSDART00000104107 |
zgc:173544
|
zgc:173544 |
| chr4_+_9467049 | 1.08 |
ENSDART00000012659
|
zgc:55888
|
zgc:55888 |
| chr10_-_34915886 | 1.05 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr8_-_16464453 | 1.01 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr25_+_3759553 | 0.98 |
ENSDART00000180601
ENSDART00000055845 ENSDART00000157050 ENSDART00000153905 |
thoc5
|
THO complex 5 |
| chr10_+_16036246 | 0.97 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr10_-_34916208 | 0.95 |
ENSDART00000187371
|
ccna1
|
cyclin A1 |
| chr1_+_30723380 | 0.94 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr19_-_4793263 | 0.92 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr16_+_47207691 | 0.91 |
ENSDART00000062507
|
ica1
|
islet cell autoantigen 1 |
| chr1_+_30723677 | 0.87 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr24_-_33366188 | 0.87 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr16_+_39159752 | 0.86 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr5_+_25733774 | 0.84 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr7_+_22313533 | 0.80 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr13_-_35808904 | 0.80 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr5_-_67241633 | 0.80 |
ENSDART00000114783
|
clip1a
|
CAP-GLY domain containing linker protein 1a |
| chr3_+_26813058 | 0.75 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr21_+_15592426 | 0.73 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr8_-_15129573 | 0.73 |
ENSDART00000142358
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr6_-_40778294 | 0.72 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr2_+_6253246 | 0.72 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr10_+_7709724 | 0.68 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr21_-_39177564 | 0.67 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr19_-_4785734 | 0.66 |
ENSDART00000113088
|
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr22_-_26274177 | 0.66 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr5_-_40292299 | 0.66 |
ENSDART00000097525
|
snx18a
|
sorting nexin 18a |
| chr21_-_2042037 | 0.66 |
ENSDART00000171131
ENSDART00000160144 |
add1
|
adducin 1 (alpha) |
| chr25_-_6049339 | 0.65 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr6_+_40922572 | 0.64 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr8_+_11325310 | 0.64 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr20_+_23947004 | 0.64 |
ENSDART00000144195
|
casp8ap2
|
caspase 8 associated protein 2 |
| chr4_-_9191220 | 0.64 |
ENSDART00000156919
|
hcfc2
|
host cell factor C2 |
| chr2_+_24936766 | 0.64 |
ENSDART00000025962
|
gyg1a
|
glycogenin 1a |
| chr20_+_19006703 | 0.63 |
ENSDART00000128435
|
pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr5_+_13394543 | 0.63 |
ENSDART00000051669
ENSDART00000135921 |
tctn2
|
tectonic family member 2 |
| chr20_-_51831816 | 0.62 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr13_-_9213207 | 0.62 |
ENSDART00000139861
ENSDART00000140524 |
HTRA2 (1 of many)
|
si:dkey-33c12.11 |
| chr25_-_2723682 | 0.62 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr22_-_21897203 | 0.61 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr5_-_30074332 | 0.60 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr16_-_54498109 | 0.60 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr3_-_30885250 | 0.59 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr25_-_2723902 | 0.59 |
ENSDART00000143721
ENSDART00000175224 |
adpgk
|
ADP-dependent glucokinase |
| chr6_-_40922971 | 0.58 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr12_+_22580579 | 0.57 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr4_+_2637947 | 0.57 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr4_-_837768 | 0.57 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr13_-_25198025 | 0.57 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr10_+_34001444 | 0.56 |
ENSDART00000149934
|
kl
|
klotho |
| chr13_+_38817871 | 0.55 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr24_+_11908833 | 0.55 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr7_-_33868903 | 0.54 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr11_+_5588122 | 0.54 |
ENSDART00000113281
|
zgc:172302
|
zgc:172302 |
| chr13_-_9236905 | 0.53 |
ENSDART00000142597
ENSDART00000147176 ENSDART00000181670 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-33c12.12 si:dkey-112g5.12 |
| chr22_-_4439311 | 0.53 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr24_+_19415124 | 0.52 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr19_-_12648122 | 0.52 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr19_+_9231766 | 0.52 |
ENSDART00000190617
|
kmt2ba
|
lysine (K)-specific methyltransferase 2Ba |
| chr2_-_40890004 | 0.51 |
ENSDART00000191746
|
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr12_+_36428052 | 0.51 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr3_-_40836081 | 0.50 |
ENSDART00000143135
|
wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr2_+_2957515 | 0.50 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr4_-_2637689 | 0.49 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr9_-_12886108 | 0.49 |
ENSDART00000177283
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr21_+_4410733 | 0.48 |
ENSDART00000136831
ENSDART00000131612 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr7_+_56703254 | 0.47 |
ENSDART00000184547
ENSDART00000004964 ENSDART00000147259 ENSDART00000134173 |
nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr11_-_11791718 | 0.47 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr5_-_22052852 | 0.47 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr22_-_37738203 | 0.46 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr24_-_25004553 | 0.46 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr2_-_1622641 | 0.46 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr12_+_47446158 | 0.45 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr13_-_24825691 | 0.45 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr20_-_37813863 | 0.44 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr24_+_11908480 | 0.44 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr12_+_3571770 | 0.44 |
ENSDART00000164707
ENSDART00000189819 |
coa3a
|
cytochrome C oxidase assembly factor 3a |
| chr14_+_21820034 | 0.44 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr22_-_20403194 | 0.44 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr1_+_13930625 | 0.43 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr14_-_14659023 | 0.43 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr12_+_1469090 | 0.43 |
ENSDART00000183637
|
usp22
|
ubiquitin specific peptidase 22 |
| chr12_+_5048044 | 0.42 |
ENSDART00000161548
ENSDART00000172607 |
kif22
|
kinesin family member 22 |
| chr13_-_9257728 | 0.42 |
ENSDART00000145985
ENSDART00000169991 ENSDART00000132764 |
HTRA2 (1 of many)
|
zgc:173425 |
| chr2_-_37462462 | 0.42 |
ENSDART00000145896
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr23_-_14766902 | 0.41 |
ENSDART00000168113
|
gss
|
glutathione synthetase |
| chr13_+_9521629 | 0.41 |
ENSDART00000149870
ENSDART00000137666 |
si:dkey-19p15.4
|
si:dkey-19p15.4 |
| chr6_+_7414215 | 0.41 |
ENSDART00000049339
|
sox21a
|
SRY (sex determining region Y)-box 21a |
| chr7_+_38680034 | 0.41 |
ENSDART00000171691
ENSDART00000007913 |
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr19_-_3724605 | 0.41 |
ENSDART00000123757
|
smim13
|
small integral membrane protein 13 |
| chr4_-_77260727 | 0.40 |
ENSDART00000075770
|
zgc:162948
|
zgc:162948 |
| chr3_-_3366590 | 0.40 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr6_+_40951227 | 0.40 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr20_-_29532939 | 0.40 |
ENSDART00000049224
ENSDART00000062377 |
taf1b
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, B |
| chr21_+_4328348 | 0.39 |
ENSDART00000131658
ENSDART00000143799 |
HTRA2 (1 of many)
|
si:dkey-84o3.7 |
| chr9_+_16241656 | 0.39 |
ENSDART00000154326
|
si:ch211-261p9.4
|
si:ch211-261p9.4 |
| chr5_-_9625459 | 0.39 |
ENSDART00000143347
|
sh2b3
|
SH2B adaptor protein 3 |
| chr4_-_73756673 | 0.39 |
ENSDART00000174274
ENSDART00000192913 ENSDART00000113546 |
BX855614.4
si:dkey-262g12.14
zgc:171551
|
si:dkey-262g12.14 zgc:171551 |
| chr11_+_42600731 | 0.38 |
ENSDART00000182753
ENSDART00000192028 ENSDART00000085868 |
appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 |
| chr19_+_12406583 | 0.38 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr9_+_43799829 | 0.37 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr8_-_50888806 | 0.37 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr13_-_9111927 | 0.37 |
ENSDART00000133815
ENSDART00000109783 ENSDART00000142540 ENSDART00000192613 |
HTRA2 (1 of many)
si:dkey-112g5.12
|
si:dkey-19p15.4 si:dkey-112g5.12 |
| chr9_+_18829360 | 0.37 |
ENSDART00000006514
|
gtf2f2b
|
general transcription factor IIF, polypeptide 2b |
| chr19_-_12648408 | 0.37 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr9_-_12595003 | 0.37 |
ENSDART00000184900
|
tra2b
|
transformer 2 beta homolog |
| chr7_+_69459759 | 0.36 |
ENSDART00000160500
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr19_-_20430892 | 0.36 |
ENSDART00000111409
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr16_-_28658341 | 0.35 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr5_-_25733745 | 0.35 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr19_-_46088429 | 0.35 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr19_-_5805923 | 0.35 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr16_+_33144306 | 0.35 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr17_+_38030327 | 0.35 |
ENSDART00000085481
|
slc25a21
|
solute carrier family 25 (mitochondrial oxoadipate carrier), member 21 |
| chr13_-_9159852 | 0.34 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr3_+_13848226 | 0.34 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr5_+_61508418 | 0.34 |
ENSDART00000023676
|
bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr13_+_9468535 | 0.33 |
ENSDART00000135088
ENSDART00000164270 ENSDART00000099619 ENSDART00000164656 |
HTRA2 (1 of many)
|
si:dkey-265c15.6 |
| chr24_-_18809433 | 0.32 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr3_+_46559639 | 0.32 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr10_-_105100 | 0.32 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr5_+_29715040 | 0.31 |
ENSDART00000192563
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr19_+_1673599 | 0.31 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr11_+_2855430 | 0.31 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr9_-_12594759 | 0.31 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr5_+_29714786 | 0.30 |
ENSDART00000148314
|
ddx31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr12_-_4408828 | 0.30 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr13_-_9184132 | 0.30 |
ENSDART00000135289
ENSDART00000136309 ENSDART00000132630 |
HTRA2 (1 of many)
|
si:dkey-33c12.10 |
| chr5_+_20257225 | 0.30 |
ENSDART00000127919
|
ssh1a
|
slingshot protein phosphatase 1a |
| chr23_+_38952529 | 0.30 |
ENSDART00000190076
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr4_+_77957611 | 0.29 |
ENSDART00000156692
|
arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr14_+_7902374 | 0.29 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr21_+_4429001 | 0.29 |
ENSDART00000134520
|
HTRA2 (1 of many)
|
si:dkey-84o3.6 |
| chr22_-_29083070 | 0.29 |
ENSDART00000104812
ENSDART00000172576 |
cbx6a
|
chromobox homolog 6a |
| chr8_-_39822917 | 0.29 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr15_-_9593532 | 0.29 |
ENSDART00000169912
|
gab2
|
GRB2-associated binding protein 2 |
| chr3_-_5067585 | 0.29 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr13_-_9070754 | 0.28 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr10_-_32494499 | 0.28 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr21_-_13662237 | 0.28 |
ENSDART00000091647
ENSDART00000151547 |
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr10_+_34175476 | 0.27 |
ENSDART00000102123
|
zgc:174193
|
zgc:174193 |
| chr10_-_3138859 | 0.27 |
ENSDART00000190606
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr22_-_7050 | 0.27 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr20_-_9123296 | 0.27 |
ENSDART00000188495
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr7_+_9308625 | 0.26 |
ENSDART00000084598
|
selenos
|
selenoprotein S |
| chr13_-_9045879 | 0.26 |
ENSDART00000155463
ENSDART00000140041 ENSDART00000137454 |
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr3_+_33345348 | 0.26 |
ENSDART00000059262
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr1_+_47335038 | 0.26 |
ENSDART00000188153
|
bcl9
|
B cell CLL/lymphoma 9 |
| chr5_-_62925282 | 0.25 |
ENSDART00000188048
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_+_36898850 | 0.25 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr21_+_4348600 | 0.25 |
ENSDART00000138463
|
HTRA2 (1 of many)
|
si:dkey-84o3.2 |
| chr9_-_3934963 | 0.24 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr16_+_33144112 | 0.24 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr10_-_3138403 | 0.24 |
ENSDART00000183365
|
ube2l3a
|
ubiquitin-conjugating enzyme E2L 3a |
| chr6_+_3334710 | 0.24 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr2_-_30721502 | 0.23 |
ENSDART00000132389
|
si:dkey-94e7.1
|
si:dkey-94e7.1 |
| chr4_+_62431132 | 0.23 |
ENSDART00000160226
|
si:ch211-79g12.1
|
si:ch211-79g12.1 |
| chr12_+_1469327 | 0.23 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr13_+_2442841 | 0.23 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr25_-_10791437 | 0.23 |
ENSDART00000127054
|
BX572619.1
|
|
| chr12_-_42214 | 0.22 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr8_+_49975160 | 0.22 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr20_+_46040666 | 0.22 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr10_-_32494304 | 0.22 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr13_-_9091354 | 0.22 |
ENSDART00000140319
ENSDART00000142697 |
HTRA2 (1 of many)
|
si:dkey-112g5.13 |
| chr22_-_22259175 | 0.22 |
ENSDART00000113824
|
hdgfl2
|
HDGF like 2 |
| chr21_-_40398932 | 0.22 |
ENSDART00000162500
|
nup98
|
nucleoporin 98 |
| chr1_+_19708508 | 0.22 |
ENSDART00000054581
ENSDART00000131206 |
march1
|
membrane-associated ring finger (C3HC4) 1 |
| chr3_-_27915270 | 0.22 |
ENSDART00000115370
|
mettl22
|
methyltransferase like 22 |
| chr24_-_32582378 | 0.21 |
ENSDART00000066590
|
rdh12l
|
retinol dehydrogenase 12, like |
| chr12_-_22238004 | 0.21 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr21_+_4313039 | 0.21 |
ENSDART00000141146
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr19_-_1871415 | 0.21 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr5_-_27993972 | 0.20 |
ENSDART00000175819
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr9_+_33145522 | 0.19 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr7_+_15308219 | 0.19 |
ENSDART00000165683
|
mespba
|
mesoderm posterior ba |
| chr8_-_45867358 | 0.19 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr19_-_2861444 | 0.18 |
ENSDART00000169053
|
clec3bb
|
C-type lectin domain family 3, member Bb |
| chr3_+_45364849 | 0.18 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr7_+_69528850 | 0.17 |
ENSDART00000109507
|
RAP1GDS1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr12_+_6098713 | 0.17 |
ENSDART00000139054
|
sgms1
|
sphingomyelin synthase 1 |
| chr14_-_47815467 | 0.17 |
ENSDART00000166827
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_+_65398161 | 0.17 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr25_+_459901 | 0.17 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr7_-_40630698 | 0.17 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr25_+_13498188 | 0.16 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr3_-_11008532 | 0.16 |
ENSDART00000165086
|
CR382337.3
|
|
| chr14_-_12020653 | 0.16 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx6-1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 3.7 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 0.5 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 1.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.8 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.8 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 1.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 1.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.7 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.4 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.4 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 0.3 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.1 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.7 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.4 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.4 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.4 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.6 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.3 | GO:0035522 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 0.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.6 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.2 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.5 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.0 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 1.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.8 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.0 | 0.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 3.2 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.2 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.5 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.8 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.5 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 1.0 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.7 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.7 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.7 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.4 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.5 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.2 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 3.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 3.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 1.6 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 1.4 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.5 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 0.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 0.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.4 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.4 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.5 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.6 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.6 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.6 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.6 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 2.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 3.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.0 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 0.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.6 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |