Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
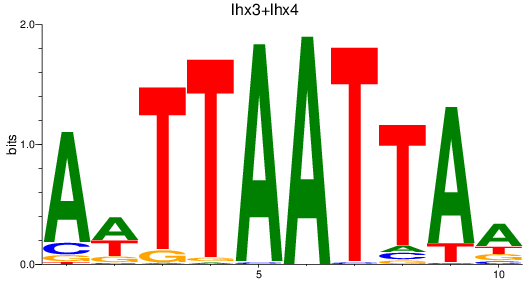
Results for lhx3+lhx4
Z-value: 1.58
Transcription factors associated with lhx3+lhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx3
|
ENSDARG00000003803 | LIM homeobox 3 |
|
lhx4
|
ENSDARG00000039458 | LIM homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| LHX3 | dr11_v1_chr5_+_71802014_71802014 | 0.87 | 3.2e-06 | Click! |
| lhx4 | dr11_v1_chr8_-_14484599_14484599 | 0.87 | 3.2e-06 | Click! |
Activity profile of lhx3+lhx4 motif
Sorted Z-values of lhx3+lhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_17083180 | 6.77 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr8_+_43340995 | 6.17 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr2_+_49799470 | 5.64 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr9_-_48407408 | 5.50 |
ENSDART00000058248
|
zgc:172182
|
zgc:172182 |
| chr24_+_35787629 | 4.67 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr18_+_25752592 | 4.58 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr7_-_28148310 | 4.47 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr5_+_69747417 | 4.42 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr8_-_13046089 | 4.41 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr5_+_6954162 | 4.34 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr15_+_21672281 | 4.31 |
ENSDART00000153923
|
si:dkey-40g16.5
|
si:dkey-40g16.5 |
| chr3_+_39853788 | 4.08 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr10_+_26972755 | 3.98 |
ENSDART00000042162
|
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr14_-_858985 | 3.97 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr11_+_25257022 | 3.86 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr1_-_34750169 | 3.77 |
ENSDART00000149380
|
si:dkey-151m22.8
|
si:dkey-151m22.8 |
| chr15_-_20939579 | 3.60 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr6_+_49551614 | 3.60 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr3_+_26145013 | 3.60 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr20_+_25586099 | 3.60 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr17_+_15433518 | 3.57 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr15_-_18162647 | 3.45 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr21_+_45502773 | 3.32 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr15_-_34567370 | 3.29 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr23_-_39666519 | 3.26 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr5_-_54672763 | 3.24 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr17_+_15433671 | 3.23 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr11_-_2838699 | 3.18 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr5_-_41494831 | 3.18 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr14_+_46313396 | 3.15 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr2_-_1548330 | 3.13 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr5_+_42386705 | 3.06 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 3.06 |
ENSDART00000188489
|
BX548073.11
|
|
| chr18_-_17485419 | 3.05 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr5_+_42400777 | 3.01 |
ENSDART00000183114
|
BX548073.8
|
|
| chr22_-_24285432 | 3.00 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr5_+_42379517 | 2.94 |
ENSDART00000103325
|
pimr59
|
Pim proto-oncogene, serine/threonine kinase, related 59 |
| chr4_+_9669717 | 2.94 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr24_-_38657683 | 2.88 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr5_+_42393896 | 2.88 |
ENSDART00000189550
|
BX548073.13
|
|
| chr17_-_39772999 | 2.87 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr17_-_24603925 | 2.83 |
ENSDART00000142589
|
si:dkey-148f10.4
|
si:dkey-148f10.4 |
| chr20_+_11731039 | 2.82 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr12_-_35830625 | 2.80 |
ENSDART00000180028
|
CU459056.1
|
|
| chr9_+_34641237 | 2.79 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr10_+_22381802 | 2.76 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr11_-_25733910 | 2.73 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr2_-_43942595 | 2.70 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr23_-_16485190 | 2.67 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr15_+_36309070 | 2.65 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr7_-_58729894 | 2.53 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr2_+_14992879 | 2.48 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr21_-_5799122 | 2.46 |
ENSDART00000129351
ENSDART00000151202 |
ccni
|
cyclin I |
| chr5_+_2815021 | 2.41 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr6_+_52350443 | 2.37 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr19_+_43297546 | 2.35 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr7_+_26629084 | 2.31 |
ENSDART00000101044
ENSDART00000173765 |
hsbp1a
|
heat shock factor binding protein 1a |
| chr20_+_4060839 | 2.27 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr4_-_9909371 | 2.24 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr14_-_1355544 | 2.23 |
ENSDART00000060417
|
cetn4
|
centrin 4 |
| chr2_+_19522082 | 2.22 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr13_-_37649595 | 2.21 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr20_+_19512727 | 2.20 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr20_-_53981626 | 2.16 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr19_+_31873308 | 2.14 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr3_-_39180048 | 2.14 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr24_+_40860320 | 2.13 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr5_-_34185115 | 2.10 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr8_-_24697586 | 2.08 |
ENSDART00000188639
|
BX324132.4
|
|
| chr14_+_6615564 | 2.00 |
ENSDART00000139292
|
si:dkeyp-44a8.2
|
si:dkeyp-44a8.2 |
| chr15_-_46779934 | 1.97 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr16_-_28856112 | 1.97 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr17_-_29119362 | 1.95 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr23_-_37113396 | 1.94 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr25_-_16146851 | 1.93 |
ENSDART00000104043
|
dkk3b
|
dickkopf WNT signaling pathway inhibitor 3b |
| chr2_-_30668580 | 1.93 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr19_-_4010263 | 1.91 |
ENSDART00000159605
ENSDART00000165541 |
map7d1b
|
MAP7 domain containing 1b |
| chr18_+_20838786 | 1.90 |
ENSDART00000138692
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr1_+_12195700 | 1.86 |
ENSDART00000040307
|
tdrd7a
|
tudor domain containing 7 a |
| chr1_-_669717 | 1.86 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr24_-_26310854 | 1.84 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr2_+_19578446 | 1.78 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr2_+_19578079 | 1.77 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr6_+_25257728 | 1.77 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr1_-_19502322 | 1.74 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr16_+_28728347 | 1.73 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr1_+_49668423 | 1.72 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr21_-_275377 | 1.70 |
ENSDART00000157509
|
rln1
|
relaxin 1 |
| chr10_+_26747755 | 1.69 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr6_-_8311044 | 1.69 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr15_-_23376541 | 1.67 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr24_+_26997798 | 1.67 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr17_+_12285285 | 1.64 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr16_-_41004731 | 1.64 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr4_-_52165969 | 1.63 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr5_-_67911111 | 1.63 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr20_-_52338782 | 1.61 |
ENSDART00000109735
ENSDART00000132941 |
si:ch1073-287p18.1
|
si:ch1073-287p18.1 |
| chr23_-_37113215 | 1.60 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr4_+_57881965 | 1.58 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr5_+_40299568 | 1.58 |
ENSDART00000142157
|
arl15a
|
ADP-ribosylation factor-like 15a |
| chr16_-_27566552 | 1.58 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr5_-_30715225 | 1.57 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr13_-_36535128 | 1.56 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr15_+_9861973 | 1.56 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr10_-_5844915 | 1.55 |
ENSDART00000185929
|
ankrd55
|
ankyrin repeat domain 55 |
| chr8_-_12403077 | 1.54 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr21_+_45841731 | 1.53 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr18_+_27821856 | 1.52 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr14_-_2933185 | 1.51 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr6_+_13117598 | 1.50 |
ENSDART00000104744
|
casp8l1
|
caspase 8, apoptosis-related cysteine peptidase, like 1 |
| chr10_-_5847655 | 1.50 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr14_-_413273 | 1.49 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr11_+_40812590 | 1.47 |
ENSDART00000186690
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| chr19_-_10043142 | 1.45 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr6_-_20952187 | 1.44 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr18_-_48547564 | 1.42 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr7_+_20475788 | 1.42 |
ENSDART00000171155
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr24_+_39108243 | 1.41 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr13_+_9046473 | 1.39 |
ENSDART00000136557
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr1_-_42289704 | 1.37 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr4_-_948776 | 1.36 |
ENSDART00000023483
|
sim1b
|
single-minded family bHLH transcription factor 1b |
| chr16_-_21140097 | 1.36 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr23_+_4253957 | 1.35 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr22_-_26865181 | 1.35 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr20_+_7584211 | 1.35 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr16_+_46111849 | 1.35 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr7_+_38811800 | 1.35 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr1_-_5455498 | 1.35 |
ENSDART00000040368
ENSDART00000114035 |
mnx2b
|
motor neuron and pancreas homeobox 2b |
| chr25_-_4146947 | 1.34 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr2_+_33368414 | 1.33 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr15_+_9327252 | 1.32 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr22_+_38173960 | 1.31 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr6_-_59942335 | 1.31 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr7_+_19552381 | 1.29 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr14_-_7137808 | 1.28 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr25_+_34915576 | 1.26 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr6_+_9427641 | 1.26 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr4_+_45148652 | 1.25 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr20_-_9095105 | 1.24 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr11_-_1509773 | 1.24 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr10_-_31015535 | 1.22 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr19_+_1688727 | 1.19 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr9_+_10692905 | 1.18 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr15_-_31514818 | 1.18 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr17_+_11675362 | 1.18 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr11_-_6048490 | 1.17 |
ENSDART00000066164
|
plvapb
|
plasmalemma vesicle associated protein b |
| chr20_+_40457599 | 1.17 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr2_+_42072231 | 1.17 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr3_-_30061985 | 1.16 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr14_+_44794936 | 1.16 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr22_-_26865361 | 1.14 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr21_-_14811058 | 1.11 |
ENSDART00000143100
|
phpt1
|
phosphohistidine phosphatase 1 |
| chr25_+_34915762 | 1.08 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr25_+_13406069 | 1.07 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr12_-_28363111 | 1.06 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr24_-_40860603 | 1.05 |
ENSDART00000188032
|
CU633479.7
|
|
| chr22_-_36530902 | 1.05 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr12_-_6880694 | 1.05 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr24_+_13316737 | 1.05 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr2_-_9059955 | 1.05 |
ENSDART00000022768
|
ak5
|
adenylate kinase 5 |
| chr9_+_28598577 | 1.05 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr15_-_21014270 | 1.04 |
ENSDART00000154019
|
si:ch211-212c13.10
|
si:ch211-212c13.10 |
| chr21_-_20939488 | 1.03 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr8_+_52637507 | 1.03 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr24_-_6078222 | 1.03 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr16_-_28878080 | 1.03 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr7_+_29954709 | 1.02 |
ENSDART00000173904
|
tpma
|
alpha-tropomyosin |
| chr18_-_15932704 | 1.02 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr25_+_7671640 | 1.02 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr22_+_16535575 | 1.01 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr22_-_22416337 | 1.01 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr3_+_46635527 | 1.00 |
ENSDART00000153971
|
si:dkey-248g21.1
|
si:dkey-248g21.1 |
| chr9_+_38481780 | 1.00 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr16_-_10053049 | 1.00 |
ENSDART00000081152
|
cep76
|
centrosomal protein 76 |
| chr7_+_72003301 | 0.98 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr22_-_5252005 | 0.98 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr14_-_46070802 | 0.98 |
ENSDART00000038670
|
elf2a
|
E74-like factor 2a (ets domain transcription factor) |
| chr18_-_43884044 | 0.97 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr22_+_19366866 | 0.97 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr23_-_18024543 | 0.97 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr15_-_14625373 | 0.96 |
ENSDART00000171841
|
slc5a2
|
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr13_+_22295905 | 0.96 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr8_+_18830759 | 0.95 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr19_-_11106315 | 0.95 |
ENSDART00000059102
|
arhgef1a
|
Rho guanine nucleotide exchange factor (GEF) 1a |
| chr22_-_31060579 | 0.94 |
ENSDART00000182376
|
cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr2_+_38373272 | 0.94 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr1_+_15204633 | 0.93 |
ENSDART00000188410
|
itln1
|
intelectin 1 |
| chr10_-_17222083 | 0.93 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr19_-_5699703 | 0.92 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr9_-_31278048 | 0.91 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr13_-_12006007 | 0.91 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr7_-_25895189 | 0.91 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr14_+_4807207 | 0.90 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr11_+_45436703 | 0.89 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr5_+_21144269 | 0.89 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr4_+_16715267 | 0.88 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr9_+_30112423 | 0.87 |
ENSDART00000112398
ENSDART00000013591 |
tfg
|
trk-fused gene |
| chr6_-_51386656 | 0.87 |
ENSDART00000154732
ENSDART00000177990 ENSDART00000184928 ENSDART00000180197 |
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr20_-_48485354 | 0.87 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr16_-_45178430 | 0.86 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx3+lhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 1.5 | 4.4 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.9 | 3.6 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.7 | 4.0 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.6 | 2.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.5 | 2.7 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.5 | 3.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.5 | 5.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.4 | 1.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.4 | 1.7 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.4 | 1.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.4 | 2.5 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.4 | 2.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.4 | 3.1 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.4 | 2.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.4 | 1.1 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.3 | 1.0 | GO:0045601 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 0.9 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.3 | 1.2 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.3 | 1.2 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.3 | 2.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.3 | 3.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.3 | 2.8 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 2.0 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 4.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.2 | 0.9 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 0.9 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.2 | 4.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.2 | 1.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 1.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) negative regulation of mesoderm development(GO:2000381) |
| 0.2 | 2.7 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 0.6 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.6 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 0.7 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.2 | 0.9 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 0.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 1.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.2 | 1.9 | GO:0030719 | P granule organization(GO:0030719) |
| 0.2 | 0.7 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 5.0 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.2 | 0.6 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 1.4 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 1.4 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.7 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 3.9 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 2.0 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.7 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.9 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 1.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 4.2 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.1 | 1.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.5 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.9 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.1 | 3.0 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 1.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.9 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.5 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.3 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.5 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.6 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 1.1 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.0 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.7 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.2 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.1 | 0.5 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 0.6 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 1.0 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 3.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 1.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.5 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 5.6 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.1 | 3.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 2.0 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.1 | 1.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 0.4 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.6 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 0.6 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 3.2 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 19.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 2.1 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.2 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.1 | 0.7 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.7 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 2.4 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.4 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 1.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 1.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 5.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.6 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 3.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.3 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 1.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 3.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 1.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.3 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 1.1 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.1 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 1.4 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.4 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.1 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.9 | 3.6 | GO:0031673 | H zone(GO:0031673) |
| 0.9 | 3.4 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 4.0 | GO:0005903 | brush border(GO:0005903) |
| 0.2 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 4.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 2.7 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 4.4 | GO:0044447 | axoneme part(GO:0044447) |
| 0.1 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 4.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 4.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 3.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 1.1 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 6.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 4.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.6 | GO:0071914 | prominosome(GO:0071914) |
| 0.1 | 0.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.9 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 2.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 3.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 4.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.9 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 1.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.5 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.8 | 2.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.7 | 4.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.6 | 4.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.5 | 3.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.5 | 6.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.4 | 4.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.4 | 1.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.4 | 2.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.4 | 1.9 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 1.1 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.2 | 0.9 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 1.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 3.6 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 2.7 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 0.7 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 2.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 2.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 0.7 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.2 | 1.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 4.0 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.6 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.7 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.9 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 1.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.4 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.8 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 1.0 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.1 | 1.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 3.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 3.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 0.2 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 1.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 5.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.5 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 3.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 3.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 3.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 1.2 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.7 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 3.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.2 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 1.0 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 14.3 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 4.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 3.6 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 3.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.2 | 0.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 1.9 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.1 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME TRANSPORT OF GLUCOSE AND OTHER SUGARS BILE SALTS AND ORGANIC ACIDS METAL IONS AND AMINE COMPOUNDS | Genes involved in Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds |
| 0.0 | 0.6 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.0 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.9 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |