Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
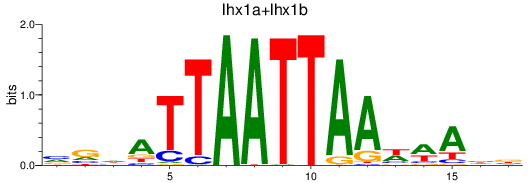
Results for lhx1a+lhx1b
Z-value: 0.57
Transcription factors associated with lhx1a+lhx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
lhx1b
|
ENSDARG00000007944 | LIM homeobox 1b |
|
lhx1a
|
ENSDARG00000014018 | LIM homeobox 1a |
|
lhx1b
|
ENSDARG00000111635 | LIM homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| lhx1a | dr11_v1_chr15_-_27710513_27710575 | 0.70 | 1.1e-03 | Click! |
| lhx1b | dr11_v1_chr5_+_56268436_56268436 | 0.68 | 1.7e-03 | Click! |
Activity profile of lhx1a+lhx1b motif
Sorted Z-values of lhx1a+lhx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_38040407 | 1.72 |
ENSDART00000139936
|
si:dkey-111e8.4
|
si:dkey-111e8.4 |
| chr3_+_39853788 | 1.66 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr18_+_46382484 | 1.61 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr8_+_43340995 | 1.40 |
ENSDART00000038566
|
rflna
|
refilin A |
| chr11_-_2838699 | 1.35 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr24_-_26328721 | 1.26 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr15_-_20939579 | 1.22 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr12_+_45200744 | 1.08 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr12_+_7497882 | 1.05 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr6_-_21582444 | 1.01 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr17_+_15433518 | 0.95 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr16_-_28856112 | 0.93 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr17_+_15433671 | 0.85 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr7_+_20471315 | 0.81 |
ENSDART00000173714
|
si:dkey-19b23.13
|
si:dkey-19b23.13 |
| chr22_-_24285432 | 0.79 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr11_-_25733910 | 0.79 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr19_+_43297546 | 0.78 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr3_+_45687266 | 0.76 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr13_-_9450210 | 0.73 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr20_-_45812144 | 0.73 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr7_-_52388734 | 0.70 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr1_-_669717 | 0.69 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr5_+_2815021 | 0.68 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr3_-_31619463 | 0.65 |
ENSDART00000124559
|
moto
|
minamoto |
| chr25_+_22296666 | 0.63 |
ENSDART00000138887
ENSDART00000193410 |
cyp11a2
|
cytochrome P450, family 11, subfamily A, polypeptide 2 |
| chr4_+_9669717 | 0.61 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr3_+_36424055 | 0.61 |
ENSDART00000170318
|
si:ch1073-443f11.2
|
si:ch1073-443f11.2 |
| chr12_-_35830625 | 0.59 |
ENSDART00000180028
|
CU459056.1
|
|
| chr18_-_17485419 | 0.58 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr20_+_25586099 | 0.57 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr4_-_9891874 | 0.56 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr24_-_38657683 | 0.53 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr23_-_16485190 | 0.53 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr4_+_72723304 | 0.50 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr6_+_17053650 | 0.50 |
ENSDART00000155296
|
pimr16
|
Pim proto-oncogene, serine/threonine kinase, related 16 |
| chr16_-_12173554 | 0.48 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr4_+_16885854 | 0.45 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr4_+_57881965 | 0.45 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr9_-_47472998 | 0.44 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr14_+_44794936 | 0.44 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr20_-_9095105 | 0.43 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr15_+_9861973 | 0.43 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr21_-_25801956 | 0.42 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr20_+_11731039 | 0.40 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr2_-_30668580 | 0.40 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr21_-_13972745 | 0.40 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr1_+_10318089 | 0.39 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr13_+_646700 | 0.39 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr16_-_12173399 | 0.39 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr9_+_28598577 | 0.38 |
ENSDART00000142623
ENSDART00000135947 |
si:ch73-7i4.1
|
si:ch73-7i4.1 |
| chr24_-_31425799 | 0.36 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr22_-_12160283 | 0.35 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr21_+_25802190 | 0.34 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr20_+_35438300 | 0.33 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr6_+_11989537 | 0.32 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr6_+_7444899 | 0.32 |
ENSDART00000053775
|
arf3b
|
ADP-ribosylation factor 3b |
| chr9_-_14504834 | 0.32 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr22_-_5252005 | 0.31 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr22_+_19552987 | 0.30 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr20_+_6035427 | 0.29 |
ENSDART00000054086
|
tshr
|
thyroid stimulating hormone receptor |
| chr8_-_45760087 | 0.29 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr12_-_6880694 | 0.28 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr7_-_38658411 | 0.28 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr9_+_34641237 | 0.27 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr2_+_38373272 | 0.26 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr2_+_42072231 | 0.26 |
ENSDART00000084517
|
vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr4_+_77943184 | 0.24 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_-_15999501 | 0.24 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr24_-_40860603 | 0.23 |
ENSDART00000188032
|
CU633479.7
|
|
| chr15_-_22074315 | 0.23 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr10_-_11261565 | 0.23 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr22_-_19552796 | 0.23 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr24_+_13316737 | 0.22 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr17_+_45405821 | 0.22 |
ENSDART00000189482
ENSDART00000177422 |
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr2_+_2223837 | 0.21 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr23_-_4409668 | 0.21 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr21_-_20939488 | 0.19 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr1_+_51039558 | 0.19 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr16_+_2820340 | 0.18 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr3_-_29941357 | 0.18 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr10_-_11261386 | 0.17 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr3_-_48716422 | 0.17 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr1_-_45616470 | 0.17 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr6_-_43677125 | 0.16 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr1_+_21731382 | 0.16 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr20_-_40755614 | 0.16 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr2_-_16217344 | 0.15 |
ENSDART00000152031
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr14_-_413273 | 0.15 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr1_+_6172786 | 0.14 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr10_+_16584382 | 0.13 |
ENSDART00000112039
|
CR790388.1
|
|
| chr11_-_41853874 | 0.13 |
ENSDART00000002556
|
mrto4
|
MRT4 homolog, ribosome maturation factor |
| chr22_+_19553390 | 0.13 |
ENSDART00000061739
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr3_-_32818607 | 0.13 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr7_+_22809905 | 0.13 |
ENSDART00000166900
ENSDART00000143455 ENSDART00000126037 |
sf1
|
splicing factor 1 |
| chr10_+_42423318 | 0.12 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr4_-_2219705 | 0.12 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr23_-_1660708 | 0.11 |
ENSDART00000175138
|
CU693481.1
|
|
| chr22_-_8725768 | 0.11 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr9_-_9415000 | 0.10 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr6_-_30839763 | 0.10 |
ENSDART00000154228
|
sgip1a
|
SH3-domain GRB2-like (endophilin) interacting protein 1a |
| chr2_+_20406399 | 0.10 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr5_-_38094130 | 0.10 |
ENSDART00000131831
|
si:ch211-284e13.4
|
si:ch211-284e13.4 |
| chr3_-_19368435 | 0.09 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr21_-_35419486 | 0.09 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr3_-_56541723 | 0.09 |
ENSDART00000156398
ENSDART00000050576 ENSDART00000184874 |
si:ch211-189a21.1
cyth1a
|
si:ch211-189a21.1 cytohesin 1a |
| chr22_-_36530902 | 0.09 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr9_+_37152564 | 0.08 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr4_+_52212057 | 0.08 |
ENSDART00000165627
|
znf1034
|
zinc finger protein 1034 |
| chr8_-_53044300 | 0.08 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr15_-_14552101 | 0.08 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr18_+_8320165 | 0.07 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr6_+_39905021 | 0.07 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr7_+_27317174 | 0.07 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr22_-_28226948 | 0.07 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr21_-_39327223 | 0.07 |
ENSDART00000115097
|
aifm5
|
apoptosis-inducing factor, mitochondrion-associated, 5 |
| chr4_-_1801519 | 0.05 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr12_+_13118540 | 0.05 |
ENSDART00000077840
ENSDART00000127870 |
cmn
|
calymmin |
| chr24_-_35767501 | 0.05 |
ENSDART00000105680
ENSDART00000042290 ENSDART00000166264 |
dtna
|
dystrobrevin, alpha |
| chr24_+_28953089 | 0.05 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr13_-_12602920 | 0.04 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr23_-_31969786 | 0.04 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr12_-_48006835 | 0.04 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr17_-_10122204 | 0.04 |
ENSDART00000160751
|
BX088587.1
|
|
| chr25_-_13490744 | 0.04 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr12_+_16168342 | 0.04 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr16_+_23303859 | 0.03 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr12_-_17712393 | 0.03 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr23_+_4689626 | 0.03 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr16_-_16761164 | 0.02 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr8_-_53926228 | 0.01 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr25_+_4751879 | 0.01 |
ENSDART00000169465
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr1_-_21287724 | 0.01 |
ENSDART00000193900
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr21_+_42717424 | 0.01 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr19_+_2631565 | 0.01 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr6_-_35052388 | 0.01 |
ENSDART00000181000
ENSDART00000170116 |
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr3_+_3681116 | 0.01 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr5_-_30620625 | 0.00 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr8_-_12867434 | 0.00 |
ENSDART00000081657
|
slc2a6
|
solute carrier family 2 (facilitated glucose transporter), member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of lhx1a+lhx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 1.6 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.3 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.8 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.4 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 1.6 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 1.3 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 1.1 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 1.2 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0070836 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.6 | GO:0042181 | ketone biosynthetic process(GO:0042181) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.3 | GO:0045762 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.1 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.0 | 0.2 | GO:0060118 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.0 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.7 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 1.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.7 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 1.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.8 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.0 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 1.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |