Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
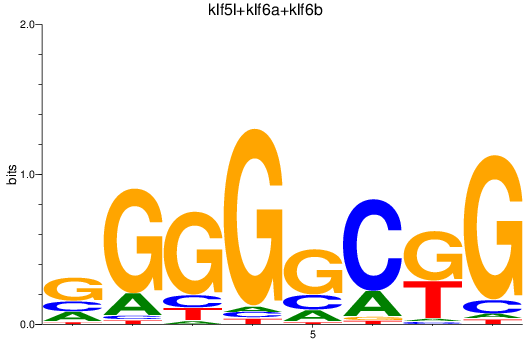
Results for klf5l+klf6a+klf6b
Z-value: 0.28
Transcription factors associated with klf5l+klf6a+klf6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf5l
|
ENSDARG00000018757 | Kruppel-like factor 5 like |
|
klf6a
|
ENSDARG00000029072 | Kruppel-like factor 6a |
|
klf6b
|
ENSDARG00000038561 | Kruppel-like factor 6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf5l | dr11_v1_chr21_-_38153824_38153824 | 0.60 | 8.8e-03 | Click! |
| klf6b | dr11_v1_chr2_+_48073972_48073972 | -0.12 | 6.3e-01 | Click! |
| klf6a | dr11_v1_chr24_-_4148914_4148915 | -0.07 | 7.9e-01 | Click! |
Activity profile of klf5l+klf6a+klf6b motif
Sorted Z-values of klf5l+klf6a+klf6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_127558 | 1.09 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr17_-_2584423 | 0.95 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr21_-_30293224 | 0.72 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr15_-_45538773 | 0.54 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr18_-_127873 | 0.49 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr20_-_49889111 | 0.45 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr3_-_21118969 | 0.41 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr2_+_59015878 | 0.38 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr17_+_6793001 | 0.35 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr16_-_7793457 | 0.35 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr6_-_7109063 | 0.33 |
ENSDART00000148548
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr20_+_26940178 | 0.33 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr13_-_12021566 | 0.32 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr7_+_25126629 | 0.32 |
ENSDART00000077217
|
zgc:101765
|
zgc:101765 |
| chr19_-_20106486 | 0.30 |
ENSDART00000043924
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr20_+_18703108 | 0.28 |
ENSDART00000181188
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr11_-_12158412 | 0.26 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr4_+_15006217 | 0.25 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr19_+_24324967 | 0.25 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr11_-_11965033 | 0.24 |
ENSDART00000193683
|
abcc10
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| chr6_+_12503849 | 0.23 |
ENSDART00000149529
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr24_+_24726956 | 0.23 |
ENSDART00000144574
ENSDART00000066628 |
mtfr1
|
mitochondrial fission regulator 1 |
| chr4_+_3438510 | 0.23 |
ENSDART00000155320
|
atxn7l1
|
ataxin 7-like 1 |
| chr5_-_23800376 | 0.22 |
ENSDART00000134184
|
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr13_-_23611960 | 0.19 |
ENSDART00000146216
|
prim2
|
DNA primase subunit 2 |
| chr3_-_30909487 | 0.18 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr18_+_6641542 | 0.18 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr20_+_700616 | 0.17 |
ENSDART00000168166
|
senp6a
|
SUMO1/sentrin specific peptidase 6a |
| chr3_+_17634952 | 0.17 |
ENSDART00000191833
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr17_-_27273296 | 0.16 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr19_+_7636941 | 0.16 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr22_+_1786230 | 0.16 |
ENSDART00000169318
ENSDART00000164948 |
znf1154
|
zinc finger protein 1154 |
| chr17_+_37253706 | 0.15 |
ENSDART00000076004
|
tmem62
|
transmembrane protein 62 |
| chr20_-_7176809 | 0.15 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr9_-_34842414 | 0.15 |
ENSDART00000126348
|
BX601644.1
|
Danio rerio cytokine receptor-like factor 2 (crlf2), mRNA. |
| chr10_-_38456382 | 0.14 |
ENSDART00000182129
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr7_-_45852270 | 0.13 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr2_+_38225427 | 0.12 |
ENSDART00000141614
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr20_+_4392687 | 0.11 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr13_+_23093743 | 0.11 |
ENSDART00000148034
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr15_-_25613114 | 0.11 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr7_-_32980017 | 0.11 |
ENSDART00000113744
|
pkp3b
|
plakophilin 3b |
| chr22_-_26558166 | 0.10 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr2_+_24868010 | 0.08 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr24_+_36452927 | 0.07 |
ENSDART00000168181
|
zgc:114120
|
zgc:114120 |
| chr3_-_33941319 | 0.07 |
ENSDART00000026090
ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr2_+_24867534 | 0.06 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr1_+_59538755 | 0.06 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr21_-_24865454 | 0.05 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr21_-_24865217 | 0.05 |
ENSDART00000101136
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr4_+_14343706 | 0.05 |
ENSDART00000142845
|
prl2
|
prolactin 2 |
| chr7_-_46019756 | 0.05 |
ENSDART00000162583
|
zgc:162297
|
zgc:162297 |
| chr14_-_237130 | 0.04 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr15_+_20543770 | 0.04 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr6_-_10788065 | 0.04 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr15_-_38202630 | 0.04 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr8_+_36621598 | 0.04 |
ENSDART00000180389
|
magixb
|
MAGI family member, X-linked b |
| chr14_+_46291022 | 0.03 |
ENSDART00000074099
|
cabp2b
|
calcium binding protein 2b |
| chr20_-_1268863 | 0.03 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr22_+_30496360 | 0.03 |
ENSDART00000189145
|
BX649490.4
|
|
| chr25_+_2229301 | 0.02 |
ENSDART00000180198
|
pparab
|
peroxisome proliferator-activated receptor alpha b |
| chr4_+_53119675 | 0.02 |
ENSDART00000167470
|
si:dkey-8o9.2
|
si:dkey-8o9.2 |
| chr9_+_13999620 | 0.02 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr16_-_563235 | 0.02 |
ENSDART00000016303
|
irx2a
|
iroquois homeobox 2a |
| chr5_-_37252111 | 0.02 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr20_+_2039518 | 0.02 |
ENSDART00000043157
|
CABZ01088134.1
|
|
| chr7_-_23768234 | 0.01 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr21_+_43253538 | 0.01 |
ENSDART00000179940
ENSDART00000164806 ENSDART00000147026 |
shroom1
|
shroom family member 1 |
| chr13_+_23214100 | 0.01 |
ENSDART00000163393
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr21_+_30293564 | 0.00 |
ENSDART00000007412
|
zmat2
|
zinc finger, matrin-type 2 |
| chr9_-_3496548 | 0.00 |
ENSDART00000102876
|
cybrd1
|
cytochrome b reductase 1 |
| chr8_+_43056153 | 0.00 |
ENSDART00000186377
|
prnpa
|
prion protein a |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf5l+klf6a+klf6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.7 | GO:0040016 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.1 | 0.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.0 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.2 | GO:1990077 | primosome complex(GO:1990077) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0001096 | RNA polymerase II basal transcription factor binding(GO:0001091) TFIIF-class transcription factor binding(GO:0001096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |