Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
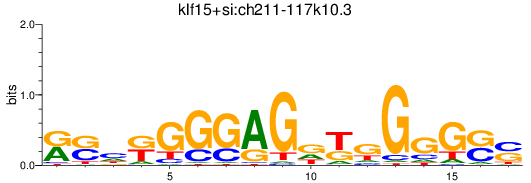
Results for klf15+si:ch211-117k10.3
Z-value: 0.43
Transcription factors associated with klf15+si:ch211-117k10.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_ch211-117k10.3
|
ENSDARG00000090914 | si_ch211-117k10.3 |
|
klf15
|
ENSDARG00000091127 | Kruppel-like factor 15 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf15 | dr11_v1_chr23_-_35195908_35195908 | -0.47 | 4.8e-02 | Click! |
| si:ch211-117k10.3 | dr11_v1_chr11_-_38513978_38513978 | 0.14 | 5.7e-01 | Click! |
Activity profile of klf15+si:ch211-117k10.3 motif
Sorted Z-values of klf15+si:ch211-117k10.3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_38110779 | 0.79 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr25_+_150570 | 0.63 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr3_-_16289826 | 0.61 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr20_-_14665002 | 0.60 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr22_+_26853254 | 0.60 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr20_-_48516765 | 0.56 |
ENSDART00000150200
|
si:zfos-223e1.2
|
si:zfos-223e1.2 |
| chr2_-_44183613 | 0.56 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr14_+_48960078 | 0.47 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr17_-_4395373 | 0.46 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr24_+_42131564 | 0.46 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr2_-_44183451 | 0.44 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr10_+_44719697 | 0.41 |
ENSDART00000158087
|
scarb1
|
scavenger receptor class B, member 1 |
| chr10_+_33382858 | 0.39 |
ENSDART00000063662
|
mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr2_-_51700709 | 0.38 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr23_+_31245395 | 0.36 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr21_-_43117327 | 0.35 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr16_+_10918252 | 0.35 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr20_-_7080427 | 0.34 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr10_-_43113731 | 0.32 |
ENSDART00000138099
|
tmem167a
|
transmembrane protein 167A |
| chr7_+_1449999 | 0.32 |
ENSDART00000173864
|
si:cabz01101003.1
|
si:cabz01101003.1 |
| chr4_-_18595525 | 0.32 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr11_+_6010177 | 0.31 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr24_-_4450238 | 0.31 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr12_-_31103906 | 0.30 |
ENSDART00000189099
ENSDART00000121527 ENSDART00000185635 ENSDART00000183754 |
tcf7l2
|
transcription factor 7 like 2 |
| chr22_-_24285432 | 0.29 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr5_-_16351306 | 0.29 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr12_+_5530247 | 0.29 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr10_+_42678520 | 0.27 |
ENSDART00000182496
|
rhobtb2b
|
Rho-related BTB domain containing 2b |
| chr16_-_12470034 | 0.26 |
ENSDART00000147483
|
ephb6
|
eph receptor B6 |
| chr14_-_9522364 | 0.26 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr2_+_7192966 | 0.23 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr14_-_32089117 | 0.23 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr16_+_11029762 | 0.23 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr14_+_22172047 | 0.22 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr11_-_40504170 | 0.22 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr8_-_34762163 | 0.22 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr10_+_39476432 | 0.21 |
ENSDART00000190375
ENSDART00000183954 |
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr16_+_9713850 | 0.21 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr1_-_38709551 | 0.21 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr17_+_53418445 | 0.20 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr1_-_39943596 | 0.20 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr22_+_26703026 | 0.20 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr4_+_3358383 | 0.19 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr7_+_23495986 | 0.19 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr12_-_15002563 | 0.18 |
ENSDART00000108852
ENSDART00000141909 |
pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr2_-_23479714 | 0.18 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr14_+_24283915 | 0.17 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr11_-_97817 | 0.16 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr13_-_45523026 | 0.15 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr8_+_1082100 | 0.15 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr6_-_442163 | 0.14 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr12_-_4540564 | 0.14 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr19_+_48117995 | 0.14 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr23_+_35759843 | 0.14 |
ENSDART00000047082
|
gdap1l1
|
ganglioside induced differentiation associated protein 1-like 1 |
| chr10_+_39476600 | 0.14 |
ENSDART00000135756
|
kirrel3a
|
kirre like nephrin family adhesion molecule 3a |
| chr18_-_26715156 | 0.14 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr18_+_33725576 | 0.14 |
ENSDART00000146816
|
si:dkey-145c18.5
|
si:dkey-145c18.5 |
| chr12_+_15002757 | 0.13 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr7_+_46261199 | 0.13 |
ENSDART00000170390
ENSDART00000183227 |
znf536
|
zinc finger protein 536 |
| chr5_-_35264517 | 0.13 |
ENSDART00000114981
|
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr5_-_30080332 | 0.13 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr23_-_40817792 | 0.12 |
ENSDART00000136343
|
si:dkeyp-27c8.1
|
si:dkeyp-27c8.1 |
| chr22_-_13042992 | 0.12 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr2_+_24781026 | 0.12 |
ENSDART00000145692
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr11_-_3334248 | 0.12 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr5_-_36916790 | 0.12 |
ENSDART00000143827
|
kptn
|
kaptin (actin binding protein) |
| chr24_+_28525507 | 0.12 |
ENSDART00000191121
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr7_-_55454406 | 0.12 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr15_-_23376541 | 0.11 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr4_-_68569527 | 0.11 |
ENSDART00000192091
|
BX548011.5
|
|
| chr3_+_23703704 | 0.11 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr24_-_40774028 | 0.10 |
ENSDART00000166578
|
CU633479.2
|
|
| chr4_-_73572030 | 0.10 |
ENSDART00000121652
|
znf1015
|
zinc finger protein 1015 |
| chr10_-_24961310 | 0.10 |
ENSDART00000037303
|
si:ch211-214k5.3
|
si:ch211-214k5.3 |
| chr9_-_8314028 | 0.10 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr18_-_26715655 | 0.10 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr17_+_33313566 | 0.10 |
ENSDART00000045040
|
si:ch211-132f19.7
|
si:ch211-132f19.7 |
| chr5_-_30074332 | 0.09 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr17_+_52822422 | 0.09 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr16_-_12914288 | 0.09 |
ENSDART00000184221
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr10_-_9115383 | 0.09 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr21_+_41649336 | 0.08 |
ENSDART00000164694
ENSDART00000181539 |
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr1_-_44638058 | 0.08 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr7_+_44445595 | 0.07 |
ENSDART00000108766
|
cdh5
|
cadherin 5 |
| chr21_+_1143141 | 0.07 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr22_-_17474583 | 0.07 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr9_+_29589790 | 0.07 |
ENSDART00000140388
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr16_-_31976269 | 0.07 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr21_+_25660613 | 0.07 |
ENSDART00000134017
|
si:dkey-17e16.15
|
si:dkey-17e16.15 |
| chr4_+_55758103 | 0.07 |
ENSDART00000185964
|
CT583728.23
|
|
| chr22_-_1168291 | 0.06 |
ENSDART00000167724
|
si:ch1073-181h11.4
|
si:ch1073-181h11.4 |
| chr4_-_11064073 | 0.06 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr20_+_35279968 | 0.06 |
ENSDART00000168216
ENSDART00000153332 |
fam49a
|
family with sequence similarity 49, member A |
| chr18_-_51015718 | 0.06 |
ENSDART00000190698
|
LO018598.1
|
|
| chr22_-_6777976 | 0.06 |
ENSDART00000187946
|
CT583625.5
|
|
| chr5_-_28606916 | 0.06 |
ENSDART00000026107
ENSDART00000137717 |
tnc
|
tenascin C |
| chr5_-_8636168 | 0.05 |
ENSDART00000134877
|
fyba
|
FYN binding protein a |
| chr6_-_43677125 | 0.05 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr22_+_7738966 | 0.05 |
ENSDART00000147073
|
si:ch73-44m9.5
|
si:ch73-44m9.5 |
| chr9_-_27442339 | 0.05 |
ENSDART00000138602
|
stxbp5l
|
syntaxin binding protein 5-like |
| chr5_+_9382301 | 0.04 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr11_-_41130239 | 0.04 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr21_-_37889727 | 0.04 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr1_-_39976492 | 0.04 |
ENSDART00000181680
|
stox2a
|
storkhead box 2a |
| chr6_+_23712911 | 0.03 |
ENSDART00000167795
|
zgc:158654
|
zgc:158654 |
| chr3_+_52467879 | 0.03 |
ENSDART00000156039
|
adgre5a
|
adhesion G protein-coupled receptor E5a |
| chr22_+_12477996 | 0.03 |
ENSDART00000177704
|
CR847870.3
|
|
| chr3_+_32425202 | 0.03 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr7_-_5711197 | 0.03 |
ENSDART00000134367
|
si:dkey-10h3.2
|
si:dkey-10h3.2 |
| chr6_+_38381957 | 0.03 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr21_-_3201027 | 0.02 |
ENSDART00000161256
|
zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr7_-_71331499 | 0.02 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr10_-_40333319 | 0.02 |
ENSDART00000150479
|
taar20a
|
trace amine associated receptor 20a |
| chr22_-_24284961 | 0.02 |
ENSDART00000141230
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr18_-_19350792 | 0.02 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr21_-_45086170 | 0.01 |
ENSDART00000188963
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr12_-_16990896 | 0.01 |
ENSDART00000152402
|
ifit12
|
interferon-induced protein with tetratricopeptide repeats 12 |
| chr3_-_36690348 | 0.01 |
ENSDART00000192513
|
myh11b
|
myosin, heavy chain 11b, smooth muscle |
| chr10_+_5645887 | 0.01 |
ENSDART00000171426
|
pdzph1
|
PDZ and pleckstrin homology domains 1 |
| chr21_+_11244068 | 0.00 |
ENSDART00000163432
|
arid6
|
AT-rich interaction domain 6 |
| chr23_+_7518294 | 0.00 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr5_+_27525477 | 0.00 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr2_+_43469241 | 0.00 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr15_-_18223769 | 0.00 |
ENSDART00000139966
|
or131-1
|
odorant receptor, family H, subfamily 131, member 1 |
| chr6_+_52918537 | 0.00 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr7_-_71331690 | 0.00 |
ENSDART00000149682
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf15+si:ch211-117k10.3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.3 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.1 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.2 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0010481 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.2 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) regulation of establishment of cell polarity(GO:2000114) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 0.6 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 0.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.6 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.2 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |