Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
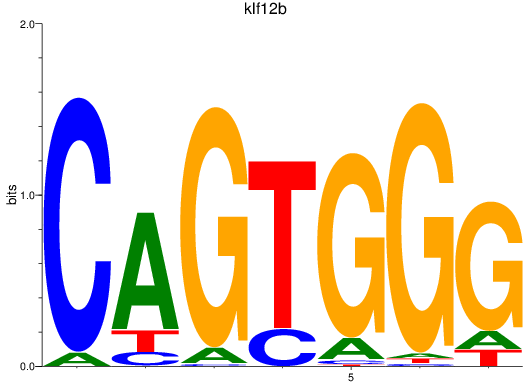
Results for klf12b
Z-value: 0.35
Transcription factors associated with klf12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12b
|
ENSDARG00000032197 | Kruppel-like factor 12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12b | dr11_v1_chr9_+_30720048_30720048 | 0.71 | 1.1e-03 | Click! |
Activity profile of klf12b motif
Sorted Z-values of klf12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_4010263 | 0.76 |
ENSDART00000159605
ENSDART00000165541 |
map7d1b
|
MAP7 domain containing 1b |
| chr3_+_17910569 | 0.71 |
ENSDART00000080946
|
ttc25
|
tetratricopeptide repeat domain 25 |
| chr22_-_20761759 | 0.70 |
ENSDART00000013803
|
amh
|
anti-Mullerian hormone |
| chr19_+_37458610 | 0.51 |
ENSDART00000103151
|
dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr2_-_29485408 | 0.50 |
ENSDART00000013411
|
cahz
|
carbonic anhydrase |
| chr22_-_2886937 | 0.49 |
ENSDART00000063533
|
aqp12
|
aquaporin 12 |
| chr15_-_20916251 | 0.48 |
ENSDART00000134053
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr1_-_52498146 | 0.47 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr22_+_16446052 | 0.47 |
ENSDART00000142454
|
si:dkey-121a11.3
|
si:dkey-121a11.3 |
| chr23_-_23401305 | 0.47 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr17_-_52643970 | 0.46 |
ENSDART00000190594
|
spred1
|
sprouty-related, EVH1 domain containing 1 |
| chr24_+_35827766 | 0.46 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr22_-_36514923 | 0.45 |
ENSDART00000187018
|
CABZ01045212.1
|
|
| chr3_-_22829710 | 0.45 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr3_+_24459709 | 0.44 |
ENSDART00000180976
|
cbx6b
|
chromobox homolog 6b |
| chr5_+_42402536 | 0.44 |
ENSDART00000186754
|
BX548073.11
|
|
| chr16_-_35579200 | 0.43 |
ENSDART00000162172
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr23_-_39636195 | 0.43 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr5_+_41774366 | 0.42 |
ENSDART00000147299
ENSDART00000083665 |
wdr66
|
WD repeat domain 66 |
| chr20_+_1412193 | 0.40 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr1_-_59116617 | 0.40 |
ENSDART00000137471
ENSDART00000140490 |
MFAP4 (1 of many)
|
si:zfos-2330d3.7 |
| chr17_-_10315644 | 0.39 |
ENSDART00000168572
|
ttc6
|
tetratricopeptide repeat domain 6 |
| chr17_-_24684687 | 0.39 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr25_+_30298377 | 0.39 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr14_-_15171435 | 0.39 |
ENSDART00000159148
ENSDART00000166622 |
si:dkey-77g12.1
|
si:dkey-77g12.1 |
| chr10_+_22381802 | 0.39 |
ENSDART00000112484
|
nlgn2b
|
neuroligin 2b |
| chr23_+_8797143 | 0.38 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr8_-_6918721 | 0.38 |
ENSDART00000014915
|
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr3_-_45822914 | 0.37 |
ENSDART00000155879
|
si:ch211-66h3.4
|
si:ch211-66h3.4 |
| chr8_-_6918290 | 0.37 |
ENSDART00000138259
ENSDART00000142496 |
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr16_-_12809873 | 0.37 |
ENSDART00000146997
ENSDART00000178291 ENSDART00000007842 |
isoc2
|
isochorismatase domain containing 2 |
| chr17_+_15433518 | 0.36 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr20_-_1378514 | 0.35 |
ENSDART00000181830
|
scara5
|
scavenger receptor class A, member 5 (putative) |
| chr16_+_5926520 | 0.34 |
ENSDART00000162229
|
ulk4
|
unc-51 like kinase 4 |
| chr3_+_28953274 | 0.34 |
ENSDART00000133528
ENSDART00000103602 |
lgals2a
|
lectin, galactoside-binding, soluble, 2a |
| chr5_-_25174420 | 0.33 |
ENSDART00000141554
|
abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2 |
| chr22_+_18530395 | 0.33 |
ENSDART00000105415
ENSDART00000183958 |
si:ch211-212d10.1
|
si:ch211-212d10.1 |
| chr8_-_12468744 | 0.33 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr2_+_22694382 | 0.32 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr14_-_36863432 | 0.32 |
ENSDART00000158052
|
rnf130
|
ring finger protein 130 |
| chr23_-_40814080 | 0.32 |
ENSDART00000135713
|
si:dkeyp-27c8.1
|
si:dkeyp-27c8.1 |
| chr21_+_25187210 | 0.32 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr10_+_439692 | 0.32 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr5_+_19867794 | 0.31 |
ENSDART00000051614
|
tchp
|
trichoplein, keratin filament binding |
| chr25_-_4146947 | 0.31 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr14_+_39255437 | 0.31 |
ENSDART00000147443
|
diaph2
|
diaphanous-related formin 2 |
| chr13_-_36535128 | 0.31 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr4_-_68563862 | 0.30 |
ENSDART00000182970
|
BX548011.2
|
|
| chr1_-_56080112 | 0.30 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr20_+_27331008 | 0.30 |
ENSDART00000141486
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr6_+_30689239 | 0.29 |
ENSDART00000065206
|
wdr78
|
WD repeat domain 78 |
| chr12_-_212843 | 0.29 |
ENSDART00000083574
|
CABZ01102039.1
|
|
| chr19_+_17400283 | 0.29 |
ENSDART00000127353
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr10_+_10351685 | 0.29 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr20_+_17739923 | 0.28 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr13_-_18105278 | 0.28 |
ENSDART00000090475
|
washc2c
|
WASH complex subunit 2C |
| chr15_-_20024205 | 0.28 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr19_-_10043142 | 0.27 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr15_-_4969525 | 0.27 |
ENSDART00000157223
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr8_+_26565512 | 0.26 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr6_+_24817852 | 0.26 |
ENSDART00000165609
|
barhl2
|
BarH-like homeobox 2 |
| chr8_-_18042089 | 0.26 |
ENSDART00000132596
|
dio1
|
iodothyronine deiodinase 1 |
| chr7_+_44713135 | 0.26 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr1_-_59139599 | 0.25 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr10_+_5159475 | 0.25 |
ENSDART00000142507
|
cdc42se2
|
CDC42 small effector 2 |
| chr4_-_880415 | 0.25 |
ENSDART00000149162
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr11_+_37652870 | 0.25 |
ENSDART00000129918
|
kif17
|
kinesin family member 17 |
| chr23_+_28494189 | 0.25 |
ENSDART00000146990
ENSDART00000006657 |
huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr13_+_21826096 | 0.25 |
ENSDART00000056432
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr10_+_17714866 | 0.24 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr4_+_6643421 | 0.24 |
ENSDART00000099462
|
gpr85
|
G protein-coupled receptor 85 |
| chr13_-_6081803 | 0.23 |
ENSDART00000099224
|
dld
|
deltaD |
| chr20_-_14875308 | 0.23 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr10_+_2975974 | 0.23 |
ENSDART00000147918
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr3_-_15886658 | 0.23 |
ENSDART00000113630
|
cramp1
|
cramped chromatin regulator homolog 1 |
| chr2_+_33368414 | 0.23 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr13_-_5079511 | 0.22 |
ENSDART00000110248
|
slc29a3
|
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
| chr8_+_40628926 | 0.22 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr13_-_48764180 | 0.22 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr25_+_15647750 | 0.22 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr3_+_15505275 | 0.22 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr20_-_44496245 | 0.22 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr8_-_39654669 | 0.22 |
ENSDART00000145677
|
si:dkey-63d15.12
|
si:dkey-63d15.12 |
| chr3_-_28750495 | 0.22 |
ENSDART00000054408
|
gsg1l
|
gsg1-like |
| chr4_-_11594202 | 0.22 |
ENSDART00000002682
|
net1
|
neuroepithelial cell transforming 1 |
| chr25_-_35497055 | 0.22 |
ENSDART00000009271
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr2_-_20599315 | 0.21 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr14_+_49251331 | 0.21 |
ENSDART00000148882
|
anxa6
|
annexin A6 |
| chr20_+_539852 | 0.21 |
ENSDART00000185994
|
dse
|
dermatan sulfate epimerase |
| chr1_-_59141715 | 0.21 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr9_-_21067971 | 0.21 |
ENSDART00000004333
|
tbx15
|
T-box 15 |
| chr5_-_38155005 | 0.20 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr3_+_52475058 | 0.20 |
ENSDART00000035867
|
si:ch211-241f5.3
|
si:ch211-241f5.3 |
| chr12_+_3301149 | 0.20 |
ENSDART00000010569
|
g6pc3
|
glucose 6 phosphatase, catalytic, 3 |
| chr10_-_27196093 | 0.20 |
ENSDART00000185282
|
auts2a
|
autism susceptibility candidate 2a |
| chr20_-_19422496 | 0.20 |
ENSDART00000143658
|
si:ch211-278j3.3
|
si:ch211-278j3.3 |
| chr2_-_16063489 | 0.20 |
ENSDART00000142828
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr6_+_50451337 | 0.20 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr21_-_24632778 | 0.20 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr16_+_5597600 | 0.19 |
ENSDART00000017307
|
zgc:91890
|
zgc:91890 |
| chr25_-_31423493 | 0.19 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr13_+_21826369 | 0.19 |
ENSDART00000165150
ENSDART00000192115 |
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr13_-_31470439 | 0.19 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr13_+_18663600 | 0.19 |
ENSDART00000141647
|
selenou1a
|
selenoprotein U 1a |
| chr1_+_17376922 | 0.19 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr3_-_2593859 | 0.19 |
ENSDART00000143826
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr25_+_5012791 | 0.19 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr19_-_26863626 | 0.18 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr5_-_22501663 | 0.18 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr23_-_12345764 | 0.18 |
ENSDART00000133956
|
phactr3a
|
phosphatase and actin regulator 3a |
| chr17_-_24364527 | 0.18 |
ENSDART00000155192
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr20_+_6659770 | 0.18 |
ENSDART00000192135
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr22_+_7439186 | 0.18 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr25_-_4525081 | 0.18 |
ENSDART00000184347
|
pidd1
|
p53-induced death domain protein 1 |
| chr2_-_41861040 | 0.18 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr15_+_4969128 | 0.18 |
ENSDART00000062856
|
rnf169
|
ring finger protein 169 |
| chr25_+_3328487 | 0.18 |
ENSDART00000181143
|
ldhbb
|
lactate dehydrogenase Bb |
| chr8_-_1884955 | 0.17 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr15_+_15415623 | 0.17 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr16_-_12173554 | 0.17 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr1_+_59062603 | 0.17 |
ENSDART00000162232
|
pin1
|
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| chr5_-_23317477 | 0.17 |
ENSDART00000090171
|
nlgn3b
|
neuroligin 3b |
| chr11_+_44503774 | 0.17 |
ENSDART00000169295
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr15_-_15968883 | 0.17 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr11_+_45448212 | 0.17 |
ENSDART00000173341
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr25_-_13842618 | 0.17 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr20_+_18580176 | 0.17 |
ENSDART00000185310
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr13_+_21870269 | 0.17 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr5_+_57641554 | 0.16 |
ENSDART00000171309
ENSDART00000157992 ENSDART00000164742 |
cryabb
|
crystallin, alpha B, b |
| chr14_-_30704075 | 0.16 |
ENSDART00000134098
|
efemp2a
|
EGF containing fibulin extracellular matrix protein 2a |
| chr3_-_40955780 | 0.16 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr14_-_83154 | 0.16 |
ENSDART00000187097
|
CABZ01086812.1
|
|
| chr12_+_18782821 | 0.16 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr19_-_44089509 | 0.16 |
ENSDART00000189136
|
rad21b
|
RAD21 cohesin complex component b |
| chr19_+_164013 | 0.16 |
ENSDART00000167379
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr5_+_22969651 | 0.16 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr9_-_21970067 | 0.16 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr2_-_10703621 | 0.15 |
ENSDART00000005944
|
rpl5a
|
ribosomal protein L5a |
| chr13_-_5937034 | 0.15 |
ENSDART00000045292
|
spred2b
|
sprouty-related, EVH1 domain containing 2b |
| chr15_-_16183583 | 0.15 |
ENSDART00000062335
|
glod4
|
glyoxalase domain containing 4 |
| chr12_-_48960308 | 0.15 |
ENSDART00000176247
|
CABZ01112647.1
|
|
| chr2_-_51644044 | 0.15 |
ENSDART00000157899
|
dad1
|
defender against cell death 1 |
| chr11_-_29685853 | 0.15 |
ENSDART00000128468
|
nr0b1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr1_-_22851481 | 0.15 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr17_-_36896560 | 0.15 |
ENSDART00000045287
|
mapre3a
|
microtubule-associated protein, RP/EB family, member 3a |
| chr17_+_10242166 | 0.15 |
ENSDART00000170420
|
clec14a
|
C-type lectin domain containing 14A |
| chr3_+_32492467 | 0.15 |
ENSDART00000151329
|
trpm4a
|
transient receptor potential cation channel, subfamily M, member 4a |
| chr1_+_20084389 | 0.15 |
ENSDART00000140263
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr21_+_17768174 | 0.14 |
ENSDART00000141380
|
rxraa
|
retinoid X receptor, alpha a |
| chr1_-_21483832 | 0.14 |
ENSDART00000102790
|
glrba
|
glycine receptor, beta a |
| chr9_-_31278048 | 0.14 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr23_-_9859989 | 0.14 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr4_-_6809323 | 0.14 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr2_+_17181777 | 0.14 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr17_+_43908428 | 0.14 |
ENSDART00000180332
|
msh4
|
mutS homolog 4 |
| chr8_+_21383347 | 0.14 |
ENSDART00000133601
ENSDART00000127545 ENSDART00000125695 |
fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr8_+_30664077 | 0.14 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr10_-_5847655 | 0.14 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr4_-_26108053 | 0.14 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr6_-_39158953 | 0.14 |
ENSDART00000050682
|
stat2
|
signal transducer and activator of transcription 2 |
| chr16_+_28792158 | 0.14 |
ENSDART00000161525
ENSDART00000027417 |
zgc:171704
|
zgc:171704 |
| chr15_-_47607480 | 0.14 |
ENSDART00000171605
|
socs9
|
suppressor of cytokine signaling 9 |
| chr7_+_61906903 | 0.13 |
ENSDART00000108540
|
tdrd7b
|
tudor domain containing 7 b |
| chr23_-_40776046 | 0.13 |
ENSDART00000136230
|
DUPD1 (1 of many)
|
si:dkeyp-27c8.1 |
| chr3_-_39198113 | 0.13 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr6_-_30210378 | 0.13 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr4_-_23643272 | 0.13 |
ENSDART00000112301
ENSDART00000133184 |
trhde.2
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 2 |
| chr22_+_3045495 | 0.13 |
ENSDART00000164061
|
LO017843.1
|
|
| chr20_+_6142433 | 0.13 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr7_+_11197940 | 0.13 |
ENSDART00000081346
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr15_+_17074185 | 0.13 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr14_-_8890437 | 0.13 |
ENSDART00000167242
|
ATG2A
|
si:ch73-45o6.2 |
| chr21_+_29077509 | 0.13 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr5_+_25074648 | 0.13 |
ENSDART00000188951
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr18_+_9171778 | 0.12 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr24_+_17068724 | 0.12 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr1_+_46948010 | 0.12 |
ENSDART00000132843
|
si:dkey-22n8.2
|
si:dkey-22n8.2 |
| chr3_-_9458445 | 0.12 |
ENSDART00000192712
|
FO904885.1
|
|
| chr5_-_31897139 | 0.12 |
ENSDART00000191957
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr4_-_76484737 | 0.12 |
ENSDART00000183816
|
ftr51
|
finTRIM family, member 51 |
| chr10_-_43771447 | 0.12 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr4_-_18201622 | 0.12 |
ENSDART00000133509
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr25_-_15040369 | 0.12 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr23_+_35714574 | 0.12 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr19_+_27479838 | 0.11 |
ENSDART00000103922
|
atat1
|
alpha tubulin acetyltransferase 1 |
| chr20_-_14665002 | 0.11 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr16_-_27749172 | 0.11 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr15_-_2754056 | 0.11 |
ENSDART00000129380
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr4_+_33191390 | 0.11 |
ENSDART00000150429
|
si:dkey-247i3.6
|
si:dkey-247i3.6 |
| chr5_+_37785152 | 0.11 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr3_-_1938588 | 0.11 |
ENSDART00000013001
ENSDART00000186405 |
zgc:152753
|
zgc:152753 |
| chr14_-_8453192 | 0.11 |
ENSDART00000136947
|
eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr4_+_359970 | 0.11 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr23_+_22658700 | 0.11 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr22_-_26323893 | 0.11 |
ENSDART00000105099
|
capn1b
|
calpain 1, (mu/I) large subunit b |
| chr2_+_24203229 | 0.11 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr7_+_50828774 | 0.11 |
ENSDART00000182821
|
per1b
|
period circadian clock 1b |
| chr4_-_20483954 | 0.11 |
ENSDART00000187240
|
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr4_-_18454591 | 0.11 |
ENSDART00000185200
|
sco2
|
SCO2 cytochrome c oxidase assembly protein |
| chr11_-_27874116 | 0.11 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 0.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.3 | GO:0021577 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.1 | 0.3 | GO:2000376 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.1 | 0.2 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.2 | GO:0003413 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.1 | 0.2 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.2 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.1 | 0.6 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.1 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.4 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.2 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.0 | 0.2 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.1 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 0.1 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.0 | 0.1 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.0 | 0.1 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.4 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 0.1 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.2 | GO:0045738 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.2 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.1 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0072337 | modified amino acid transport(GO:0072337) |
| 0.0 | 0.1 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.1 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.0 | GO:2000793 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.5 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.0 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.9 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.0 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 0.2 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.3 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.1 | 0.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |