Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
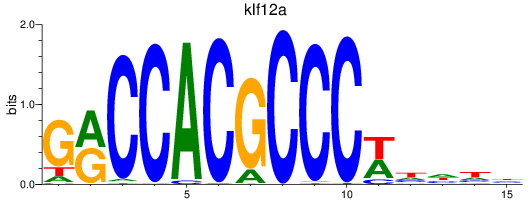
Results for klf12a
Z-value: 0.46
Transcription factors associated with klf12a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
klf12a
|
ENSDARG00000015312 | Kruppel-like factor 12a |
|
klf12a
|
ENSDARG00000115152 | Kruppel-like factor 12a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| klf12a | dr11_v1_chr1_+_34527213_34527213 | -0.23 | 3.5e-01 | Click! |
Activity profile of klf12a motif
Sorted Z-values of klf12a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_17197546 | 2.10 |
ENSDART00000139939
ENSDART00000135146 ENSDART00000063800 ENSDART00000163606 |
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr7_+_1473929 | 1.50 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr17_-_18898115 | 1.46 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr12_+_46634736 | 1.43 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr22_+_835728 | 1.40 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr8_-_32385989 | 1.35 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr13_-_6252498 | 1.31 |
ENSDART00000115157
|
tuba4l
|
tubulin, alpha 4 like |
| chr10_-_34002185 | 1.25 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr8_-_410728 | 1.25 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr2_+_59015878 | 1.06 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr5_-_69482891 | 1.06 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr23_-_31645760 | 1.05 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr20_-_182841 | 0.96 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr11_-_34577034 | 0.93 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr7_-_59159253 | 0.92 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr19_+_46113828 | 0.91 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr9_+_8380728 | 0.91 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr21_-_13856689 | 0.88 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr1_+_45707219 | 0.87 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr17_+_17804752 | 0.86 |
ENSDART00000123350
|
sptlc2a
|
serine palmitoyltransferase, long chain base subunit 2a |
| chr3_-_40054615 | 0.84 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr20_-_4031475 | 0.83 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr21_+_34167178 | 0.81 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr8_+_47219107 | 0.81 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr24_-_11076400 | 0.80 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr4_-_4612116 | 0.78 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr14_+_7048930 | 0.77 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr11_+_45287541 | 0.76 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr10_-_44017642 | 0.72 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr2_-_32513538 | 0.72 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr16_-_17713859 | 0.70 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr5_-_9824908 | 0.67 |
ENSDART00000169698
|
zgc:158343
|
zgc:158343 |
| chr23_+_30730121 | 0.64 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr22_+_24623936 | 0.63 |
ENSDART00000160924
|
mcoln2
|
mucolipin 2 |
| chr5_+_36850650 | 0.62 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr24_-_36238054 | 0.62 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr7_+_55292959 | 0.62 |
ENSDART00000147539
ENSDART00000073555 |
ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr16_+_30002605 | 0.58 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr7_+_38349667 | 0.58 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr25_+_7492663 | 0.58 |
ENSDART00000166496
|
cat
|
catalase |
| chr15_+_29025090 | 0.57 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr17_+_16429826 | 0.57 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr7_-_19364813 | 0.57 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr5_-_23696926 | 0.57 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr6_+_41503854 | 0.57 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr14_-_24110707 | 0.56 |
ENSDART00000133522
ENSDART00000123152 |
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr8_+_104114 | 0.56 |
ENSDART00000172101
|
sncaip
|
synuclein, alpha interacting protein |
| chr20_-_15104709 | 0.56 |
ENSDART00000034011
|
fmo5
|
flavin containing monooxygenase 5 |
| chr16_+_43077909 | 0.55 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr2_+_25658112 | 0.54 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr22_-_20924747 | 0.54 |
ENSDART00000185845
ENSDART00000179672 |
ell
|
elongation factor RNA polymerase II |
| chr3_-_32362872 | 0.54 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr13_+_42309688 | 0.53 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr22_-_20924564 | 0.53 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr25_+_186583 | 0.53 |
ENSDART00000161504
|
pclaf
|
PCNA clamp associated factor |
| chr11_+_13524504 | 0.52 |
ENSDART00000139889
|
arrdc2
|
arrestin domain containing 2 |
| chr17_+_35097024 | 0.51 |
ENSDART00000026152
|
asap2a
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2a |
| chr15_+_28410664 | 0.50 |
ENSDART00000132028
ENSDART00000057697 ENSDART00000057257 |
pitpnaa
|
phosphatidylinositol transfer protein, alpha a |
| chr5_-_36597612 | 0.50 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr21_-_22122312 | 0.49 |
ENSDART00000101726
|
slc35f2
|
solute carrier family 35, member F2 |
| chr21_+_1647990 | 0.48 |
ENSDART00000148540
|
fech
|
ferrochelatase |
| chr6_-_40657653 | 0.48 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr8_-_410199 | 0.47 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr6_-_7776612 | 0.47 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr18_+_38191346 | 0.46 |
ENSDART00000052703
|
nucb2b
|
nucleobindin 2b |
| chr25_+_6122823 | 0.43 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr2_-_39675829 | 0.43 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr5_-_12031174 | 0.43 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr17_-_24889837 | 0.43 |
ENSDART00000187980
|
gale
|
UDP-galactose-4-epimerase |
| chr10_+_1052591 | 0.42 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr21_-_32060993 | 0.42 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr21_+_21279159 | 0.42 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr16_-_7793457 | 0.41 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr3_-_31924643 | 0.41 |
ENSDART00000122589
|
rnf113a
|
ring finger protein 113A |
| chr7_+_55518519 | 0.41 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr6_+_7533601 | 0.41 |
ENSDART00000057823
|
pa2g4a
|
proliferation-associated 2G4, a |
| chr3_-_32541033 | 0.40 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr3_+_32410746 | 0.40 |
ENSDART00000025496
|
rras
|
RAS related |
| chr21_+_22330005 | 0.40 |
ENSDART00000140751
ENSDART00000157839 ENSDART00000140468 |
nadk2
|
NAD kinase 2, mitochondrial |
| chr24_-_26632171 | 0.40 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr15_-_19128705 | 0.40 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr4_+_9011448 | 0.40 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr5_+_43458304 | 0.40 |
ENSDART00000051114
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr15_-_1001177 | 0.40 |
ENSDART00000160730
|
zgc:162936
|
zgc:162936 |
| chr22_+_5118361 | 0.39 |
ENSDART00000168371
ENSDART00000170222 ENSDART00000158846 |
mibp
|
muscle-specific beta 1 integrin binding protein |
| chr8_-_4618653 | 0.39 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr25_+_3788443 | 0.39 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr17_+_44441042 | 0.38 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr11_+_5499661 | 0.38 |
ENSDART00000027850
|
slc35e1
|
solute carrier family 35, member E1 |
| chr9_+_50110763 | 0.38 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr2_+_35880600 | 0.37 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr20_-_51656512 | 0.37 |
ENSDART00000129965
|
LO018154.1
|
|
| chr17_+_25332711 | 0.37 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr2_+_25657958 | 0.36 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr20_-_25748407 | 0.36 |
ENSDART00000063152
|
chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr10_-_41352502 | 0.36 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr2_+_25560556 | 0.36 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr4_+_9011825 | 0.36 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr9_+_30421489 | 0.34 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr20_+_35058634 | 0.34 |
ENSDART00000122696
|
hnrnpub
|
heterogeneous nuclear ribonucleoprotein Ub |
| chr5_+_4006837 | 0.34 |
ENSDART00000138862
|
pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr2_+_11028923 | 0.34 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr23_-_18707418 | 0.34 |
ENSDART00000144668
ENSDART00000141205 ENSDART00000016765 |
zgc:103759
|
zgc:103759 |
| chr16_-_26132122 | 0.33 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr2_+_43894368 | 0.33 |
ENSDART00000143885
|
gbp3
|
guanylate binding protein 3 |
| chr22_-_21676364 | 0.33 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr13_+_28618086 | 0.33 |
ENSDART00000087001
|
cnnm2a
|
cyclin and CBS domain divalent metal cation transport mediator 2a |
| chr8_-_1698155 | 0.32 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr23_-_27822920 | 0.32 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr7_+_40083601 | 0.32 |
ENSDART00000099046
|
zgc:112356
|
zgc:112356 |
| chr3_+_15893039 | 0.32 |
ENSDART00000055780
|
jpt2
|
Jupiter microtubule associated homolog 2 |
| chr6_-_25165693 | 0.32 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr1_+_1712140 | 0.31 |
ENSDART00000081047
|
atp1a1a.1
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 1 |
| chr1_-_8653385 | 0.31 |
ENSDART00000193041
|
actb1
|
actin, beta 1 |
| chr5_+_61658282 | 0.31 |
ENSDART00000188878
|
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr20_-_13140309 | 0.31 |
ENSDART00000020703
ENSDART00000188594 |
ints7
|
integrator complex subunit 7 |
| chr23_-_10745288 | 0.31 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr3_-_52899394 | 0.30 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr6_-_1591002 | 0.30 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr13_+_421231 | 0.30 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr21_-_11657043 | 0.30 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr10_+_29204581 | 0.29 |
ENSDART00000148503
|
picalma
|
phosphatidylinositol binding clathrin assembly protein a |
| chr17_-_44440832 | 0.29 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr18_+_6558338 | 0.29 |
ENSDART00000110892
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr13_-_42673978 | 0.29 |
ENSDART00000133848
ENSDART00000099738 ENSDART00000099729 ENSDART00000169083 |
lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr12_-_48312647 | 0.29 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr16_+_53455638 | 0.29 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr10_+_37500234 | 0.28 |
ENSDART00000132096
ENSDART00000099473 |
msi2a
|
musashi RNA-binding protein 2a |
| chr19_-_31686252 | 0.28 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr5_+_61657702 | 0.28 |
ENSDART00000134387
ENSDART00000171248 |
ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr18_+_6558146 | 0.28 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr16_+_19637384 | 0.27 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr7_+_29044888 | 0.27 |
ENSDART00000086871
|
gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr16_-_31351419 | 0.27 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr25_+_35375848 | 0.27 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr23_-_5818992 | 0.27 |
ENSDART00000148730
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr3_-_40276057 | 0.26 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr20_+_29565906 | 0.26 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr10_+_6383270 | 0.26 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr6_+_23122789 | 0.26 |
ENSDART00000049226
ENSDART00000067560 |
acox1
|
acyl-CoA oxidase 1, palmitoyl |
| chr6_+_18520859 | 0.26 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr17_-_51260476 | 0.26 |
ENSDART00000084348
|
trappc12
|
trafficking protein particle complex 12 |
| chr24_-_38644937 | 0.26 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr15_-_19443997 | 0.26 |
ENSDART00000114936
|
esamb
|
endothelial cell adhesion molecule b |
| chr22_-_3172823 | 0.25 |
ENSDART00000167550
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr23_-_19831739 | 0.25 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr8_+_47099033 | 0.25 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr23_-_3759345 | 0.25 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr15_-_9593532 | 0.25 |
ENSDART00000169912
|
gab2
|
GRB2-associated binding protein 2 |
| chr16_+_26747766 | 0.24 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr22_-_38224315 | 0.24 |
ENSDART00000165430
ENSDART00000140968 |
hps3
|
Hermansky-Pudlak syndrome 3 |
| chr14_+_31566517 | 0.24 |
ENSDART00000002684
|
ints6l
|
integrator complex subunit 6 like |
| chr18_-_7481036 | 0.24 |
ENSDART00000101292
|
si:dkey-238c7.16
|
si:dkey-238c7.16 |
| chr2_+_58221163 | 0.24 |
ENSDART00000157939
|
FO704813.1
|
|
| chr20_+_1196386 | 0.24 |
ENSDART00000041192
|
lyrm2
|
LYR motif containing 2 |
| chr1_+_53714734 | 0.23 |
ENSDART00000114689
|
pus10
|
pseudouridylate synthase 10 |
| chr1_-_493218 | 0.23 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr25_-_6049339 | 0.23 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr3_+_19685873 | 0.23 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr19_+_8144556 | 0.23 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr5_+_68807170 | 0.23 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr5_-_43819663 | 0.23 |
ENSDART00000022481
|
mccc2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr3_-_50998577 | 0.23 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr17_+_6765621 | 0.22 |
ENSDART00000156637
ENSDART00000007622 |
afg1la
|
AFG1 like ATPase a |
| chr10_+_33382858 | 0.22 |
ENSDART00000063662
|
mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr19_-_34011340 | 0.22 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr13_-_44808783 | 0.22 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr18_+_20047374 | 0.22 |
ENSDART00000146957
|
uacaa
|
uveal autoantigen with coiled-coil domains and ankyrin repeats a |
| chr1_-_55058795 | 0.21 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr16_-_280835 | 0.21 |
ENSDART00000190541
|
LO017917.1
|
|
| chr1_-_55210619 | 0.21 |
ENSDART00000111671
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr11_-_25384213 | 0.20 |
ENSDART00000103650
|
mafbb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Bb |
| chr25_+_4635355 | 0.20 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr15_-_5580093 | 0.20 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr18_+_27077853 | 0.20 |
ENSDART00000125326
ENSDART00000192660 ENSDART00000098334 |
ppp1r15b
|
protein phosphatase 1, regulatory subunit 15B |
| chr1_+_45922699 | 0.20 |
ENSDART00000033669
|
lipt1
|
lipoyltransferase 1 |
| chr13_-_33671694 | 0.20 |
ENSDART00000143945
ENSDART00000100504 |
zgc:163030
|
zgc:163030 |
| chr25_-_37284370 | 0.19 |
ENSDART00000103222
|
nudt7
|
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
| chr21_-_5056812 | 0.18 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr14_-_24277805 | 0.18 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr11_+_3005536 | 0.18 |
ENSDART00000174539
|
cpne5b
|
copine Vb |
| chr19_-_2421793 | 0.18 |
ENSDART00000180238
|
TMEM196 (1 of many)
|
transmembrane protein 196 |
| chr21_-_14692119 | 0.18 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr3_+_17951790 | 0.17 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr24_-_20152965 | 0.17 |
ENSDART00000147004
|
oxsr1b
|
oxidative stress responsive 1b |
| chr17_-_25831569 | 0.17 |
ENSDART00000148743
|
hhat
|
hedgehog acyltransferase |
| chr19_+_29854223 | 0.17 |
ENSDART00000109729
ENSDART00000157419 |
si:ch73-130a3.4
|
si:ch73-130a3.4 |
| chr3_+_22984098 | 0.17 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr7_-_73815262 | 0.16 |
ENSDART00000185351
|
zgc:165555
|
zgc:165555 |
| chr8_+_20157798 | 0.16 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr19_+_7154500 | 0.16 |
ENSDART00000035967
ENSDART00000160894 |
brd2a
|
bromodomain containing 2a |
| chr22_-_2922053 | 0.16 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr13_-_36621926 | 0.16 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr23_-_17429775 | 0.16 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr9_-_3149896 | 0.16 |
ENSDART00000020861
|
pdk1
|
pyruvate dehydrogenase kinase, isozyme 1 |
| chr23_-_3759692 | 0.15 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr5_+_23136544 | 0.15 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr25_+_19008497 | 0.15 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr11_-_40647190 | 0.15 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr14_+_33264303 | 0.15 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of klf12a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:2001014 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 0.9 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 1.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.7 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 1.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.5 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.7 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 0.6 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.3 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.5 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.4 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.4 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 0.8 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.9 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.8 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.8 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.7 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.1 | 0.4 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 | 2.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.5 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.7 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.6 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.4 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.4 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.7 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.2 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.5 | GO:0043249 | heme biosynthetic process(GO:0006783) erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.0 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.3 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 1.3 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.1 | 0.9 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.3 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.3 | GO:0098888 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.4 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.3 | 0.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.4 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.9 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.3 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.6 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.2 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.3 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.3 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 0.4 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.9 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.8 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.5 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.1 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 1.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.6 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.7 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.3 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |