Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for jund_batf
Z-value: 1.23
Transcription factors associated with jund_batf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
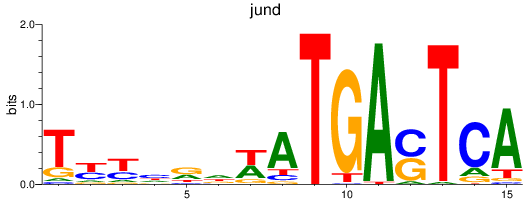
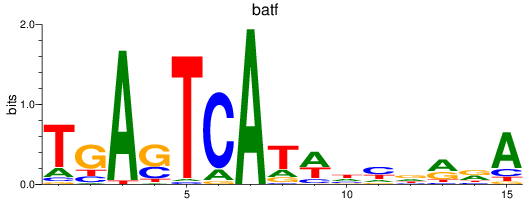
jund
|
ENSDARG00000067850 | JunD proto-oncogene, AP-1 transcription factor subunit |
|
batf
|
ENSDARG00000011818 | basic leucine zipper transcription factor, ATF-like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| batf | dr11_v1_chr20_+_46572550_46572550 | -0.96 | 2.2e-10 | Click! |
| jund | dr11_v1_chr2_-_56131312_56131312 | 0.66 | 2.7e-03 | Click! |
Activity profile of jund_batf motif
Sorted Z-values of jund_batf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_13818061 | 5.19 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr7_-_52963493 | 5.12 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr9_+_2499627 | 4.94 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr24_+_13277573 | 4.75 |
ENSDART00000137886
|
si:ch211-171b20.3
|
si:ch211-171b20.3 |
| chr24_+_31361407 | 4.61 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr6_+_7421898 | 4.38 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr3_+_24197934 | 4.02 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr21_+_22845317 | 3.99 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr20_+_2134816 | 3.97 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr22_+_16497670 | 3.97 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr24_+_35827766 | 3.93 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr22_+_36914636 | 3.80 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr25_-_15496485 | 3.48 |
ENSDART00000140245
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr3_+_24986145 | 3.48 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr5_+_13385837 | 3.36 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr1_+_135903 | 2.97 |
ENSDART00000124837
|
f10
|
coagulation factor X |
| chr12_+_1455147 | 2.84 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr20_-_2134620 | 2.73 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr19_+_37118547 | 2.60 |
ENSDART00000103163
|
cx30.9
|
connexin 30.9 |
| chr6_+_49095646 | 2.58 |
ENSDART00000103385
|
slc25a55a
|
solute carrier family 25, member 55a |
| chr8_-_36554675 | 2.46 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr3_+_31621774 | 2.14 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr3_-_26017592 | 2.09 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr4_-_69189894 | 2.07 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr20_-_32658305 | 2.07 |
ENSDART00000140622
ENSDART00000152909 |
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr15_-_20933574 | 1.99 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr16_-_50203058 | 1.89 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr16_+_42830152 | 1.88 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr22_+_38173960 | 1.75 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr3_-_16289826 | 1.70 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr8_+_25900049 | 1.69 |
ENSDART00000124300
ENSDART00000127618 ENSDART00000024009 |
rhoab
|
ras homolog gene family, member Ab |
| chr7_-_24236364 | 1.68 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr3_-_26017831 | 1.67 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr16_-_38518270 | 1.65 |
ENSDART00000131899
|
rspo2
|
R-spondin 2 |
| chr20_+_25340814 | 1.64 |
ENSDART00000063028
|
ctgfa
|
connective tissue growth factor a |
| chr11_-_2478374 | 1.60 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr25_-_15512819 | 1.48 |
ENSDART00000142684
|
si:dkeyp-67e1.2
|
si:dkeyp-67e1.2 |
| chr25_+_27873671 | 1.44 |
ENSDART00000088817
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr3_-_16055432 | 1.43 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr15_-_34558579 | 1.40 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr10_+_31344227 | 1.35 |
ENSDART00000184470
|
AL845320.2
|
|
| chr25_+_27873836 | 1.35 |
ENSDART00000163801
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr24_+_26658132 | 1.34 |
ENSDART00000183081
|
FQ378040.1
|
|
| chr17_-_33714636 | 1.33 |
ENSDART00000188500
|
DNAL1
|
si:dkey-84k17.3 |
| chr21_+_17024002 | 1.33 |
ENSDART00000080628
|
arpc3
|
actin related protein 2/3 complex, subunit 3 |
| chr25_-_24233884 | 1.32 |
ENSDART00000146419
|
si:dkey-11e23.9
|
si:dkey-11e23.9 |
| chr10_+_28428222 | 1.32 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr12_+_7497882 | 1.31 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr8_+_50531709 | 1.28 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr1_-_56176976 | 1.17 |
ENSDART00000052688
|
c3a.1
|
complement component c3a, duplicate 1 |
| chr18_-_36909773 | 1.16 |
ENSDART00000141694
|
si:ch211-160d20.5
|
si:ch211-160d20.5 |
| chr7_+_528593 | 1.15 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr12_+_19188542 | 1.12 |
ENSDART00000134726
ENSDART00000148011 ENSDART00000109541 |
cby1
|
chibby homolog 1 (Drosophila) |
| chr25_-_15504559 | 1.10 |
ENSDART00000139294
|
BX323543.5
|
|
| chr1_-_40086978 | 1.06 |
ENSDART00000136990
|
si:dkey-117m1.4
|
si:dkey-117m1.4 |
| chr13_-_20381485 | 1.04 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr18_-_5692292 | 1.02 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr21_+_33459524 | 1.02 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr6_-_31224563 | 1.01 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr10_+_42733210 | 0.98 |
ENSDART00000189832
|
CABZ01063556.1
|
|
| chr15_-_21877726 | 0.97 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr5_+_38276582 | 0.96 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr9_-_7640692 | 0.95 |
ENSDART00000135616
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr21_+_8341774 | 0.88 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr11_-_39202915 | 0.88 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr5_-_57480660 | 0.85 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr3_-_41535647 | 0.82 |
ENSDART00000153723
ENSDART00000154198 |
si:ch211-222n22.1
|
si:ch211-222n22.1 |
| chr14_-_26400501 | 0.82 |
ENSDART00000054189
|
pimr212
|
Pim proto-oncogene, serine/threonine kinase, related 212 |
| chr23_-_44848961 | 0.79 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr14_+_49382180 | 0.79 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr22_+_19365220 | 0.78 |
ENSDART00000132781
ENSDART00000135672 ENSDART00000153630 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr24_-_21404367 | 0.77 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr25_-_18330503 | 0.75 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr23_+_29883216 | 0.72 |
ENSDART00000113367
ENSDART00000149761 ENSDART00000148706 |
aurkaip1
|
aurora kinase A interacting protein 1 |
| chr5_-_30924347 | 0.70 |
ENSDART00000111749
ENSDART00000086564 ENSDART00000153909 |
spns2
|
spinster homolog 2 (Drosophila) |
| chr13_-_5257303 | 0.68 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr7_+_24496894 | 0.67 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr23_-_18024543 | 0.67 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr19_+_48102560 | 0.66 |
ENSDART00000164464
|
utp18
|
UTP18 small subunit (SSU) processome component |
| chr12_+_9551667 | 0.64 |
ENSDART00000048493
ENSDART00000166178 |
p4ha1a
|
prolyl 4-hydroxylase, alpha polypeptide I a |
| chr16_-_32184460 | 0.63 |
ENSDART00000102027
|
kpna5
|
karyopherin alpha 5 (importin alpha 6) |
| chr5_-_30984010 | 0.63 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr8_+_29856013 | 0.62 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr23_+_40109353 | 0.62 |
ENSDART00000149249
|
ghrhrl
|
growth hormone releasing hormone receptor, like |
| chr6_-_48317562 | 0.62 |
ENSDART00000103425
|
st7l
|
suppression of tumorigenicity 7 like |
| chr15_+_29123031 | 0.60 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr9_-_2572790 | 0.60 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr13_+_10621257 | 0.60 |
ENSDART00000008603
|
prepl
|
prolyl endopeptidase-like |
| chr22_+_19247255 | 0.58 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr16_+_49005321 | 0.58 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr17_-_6536305 | 0.57 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr25_+_7423770 | 0.56 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr5_+_17624463 | 0.55 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr13_-_50624743 | 0.54 |
ENSDART00000167949
|
vox
|
ventral homeobox |
| chr20_+_34868933 | 0.54 |
ENSDART00000153006
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr20_-_35040041 | 0.54 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr12_+_41348969 | 0.53 |
ENSDART00000171352
|
si:ch211-27e6.1
|
si:ch211-27e6.1 |
| chr21_+_9628854 | 0.53 |
ENSDART00000161753
ENSDART00000160711 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr14_+_48062180 | 0.53 |
ENSDART00000056713
|
ppid
|
peptidylprolyl isomerase D |
| chr10_-_40514643 | 0.52 |
ENSDART00000140705
|
taar19k
|
trace amine associated receptor 19k |
| chr2_-_6182098 | 0.52 |
ENSDART00000156167
|
si:ch73-182a11.2
|
si:ch73-182a11.2 |
| chr3_+_60044780 | 0.51 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr1_-_22834824 | 0.49 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr18_-_13360106 | 0.48 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr18_+_45114392 | 0.46 |
ENSDART00000172328
|
large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr5_-_30487822 | 0.45 |
ENSDART00000189288
ENSDART00000183201 |
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr2_-_49997055 | 0.45 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr8_+_8973425 | 0.44 |
ENSDART00000066107
|
bcap31
|
B cell receptor associated protein 31 |
| chr17_+_23770848 | 0.43 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr5_-_43959972 | 0.43 |
ENSDART00000180517
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr19_-_36675023 | 0.42 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr3_+_37707432 | 0.42 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr23_+_20863145 | 0.40 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr7_-_37563883 | 0.40 |
ENSDART00000148805
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr23_+_9067131 | 0.39 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr24_+_7800486 | 0.38 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr1_+_7517454 | 0.38 |
ENSDART00000016139
|
lancl1
|
LanC antibiotic synthetase component C-like 1 (bacterial) |
| chr4_-_72080351 | 0.38 |
ENSDART00000174925
|
LO017820.1
|
|
| chr7_-_35408618 | 0.38 |
ENSDART00000074963
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr2_-_6232517 | 0.38 |
ENSDART00000139842
|
rec8a
|
REC8 meiotic recombination protein a |
| chr19_+_33093395 | 0.36 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr12_+_21684239 | 0.36 |
ENSDART00000043546
|
mrpl27
|
mitochondrial ribosomal protein L27 |
| chr3_-_61185746 | 0.36 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr17_-_52091999 | 0.36 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr21_+_25071805 | 0.34 |
ENSDART00000078651
|
dixdc1b
|
DIX domain containing 1b |
| chr5_+_61556172 | 0.34 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr25_-_11026907 | 0.33 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr2_+_30379650 | 0.33 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr18_-_28938912 | 0.30 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr9_+_38372216 | 0.30 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr13_-_21701323 | 0.30 |
ENSDART00000164112
|
si:dkey-191g9.7
|
si:dkey-191g9.7 |
| chr4_-_76270779 | 0.30 |
ENSDART00000183709
ENSDART00000192689 |
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr5_-_41531629 | 0.30 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr20_-_42972599 | 0.29 |
ENSDART00000100751
|
pomcb
|
proopiomelanocortin b |
| chr13_-_50624173 | 0.29 |
ENSDART00000184181
|
vox
|
ventral homeobox |
| chr7_-_35409027 | 0.28 |
ENSDART00000128334
|
lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr4_+_68087932 | 0.28 |
ENSDART00000150494
|
znf1096
|
zinc finger protein 1096 |
| chr24_+_17260001 | 0.28 |
ENSDART00000066765
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr23_+_39558508 | 0.28 |
ENSDART00000017902
|
camk1gb
|
calcium/calmodulin-dependent protein kinase IGb |
| chr24_+_26039464 | 0.28 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr19_+_40856807 | 0.28 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr19_+_42142381 | 0.28 |
ENSDART00000129915
|
kcnq4
|
potassium voltage-gated channel, KQT-like subfamily, member 4 |
| chr8_+_39634114 | 0.27 |
ENSDART00000144293
|
msi1
|
musashi RNA-binding protein 1 |
| chr21_+_5888641 | 0.26 |
ENSDART00000091331
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr8_+_13389115 | 0.26 |
ENSDART00000184428
ENSDART00000154266 ENSDART00000049469 |
jak3
|
Janus kinase 3 (a protein tyrosine kinase, leukocyte) |
| chr9_+_14010823 | 0.25 |
ENSDART00000143837
|
si:ch211-67e16.3
|
si:ch211-67e16.3 |
| chr13_+_35637875 | 0.25 |
ENSDART00000180657
|
thbs2a
|
thrombospondin 2a |
| chr10_-_43928937 | 0.23 |
ENSDART00000168014
ENSDART00000086220 |
mctp1b
|
multiple C2 domains, transmembrane 1b |
| chr2_-_16224083 | 0.23 |
ENSDART00000165953
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr20_+_53474963 | 0.23 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr8_+_43053519 | 0.23 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr15_+_17446796 | 0.22 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr4_+_33654247 | 0.21 |
ENSDART00000192537
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr3_+_30968176 | 0.21 |
ENSDART00000186266
|
prf1.9
|
perforin 1.9 |
| chr14_-_17588345 | 0.21 |
ENSDART00000143486
|
selenot2
|
selenoprotein T, 2 |
| chr20_+_20726231 | 0.20 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr17_-_6536466 | 0.20 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr14_-_32884138 | 0.20 |
ENSDART00000105726
|
slc25a5
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr20_-_34127415 | 0.19 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr20_+_23390984 | 0.19 |
ENSDART00000136922
|
fryl
|
furry homolog, like |
| chr4_-_30057608 | 0.19 |
ENSDART00000184429
|
si:rp71-7l19.2
|
si:rp71-7l19.2 |
| chr14_-_976912 | 0.18 |
ENSDART00000114053
|
arsia
|
arylsulfatase family, member Ia |
| chr13_+_35339182 | 0.18 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr4_+_76957765 | 0.18 |
ENSDART00000129607
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr8_+_27807974 | 0.18 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr15_-_29586747 | 0.18 |
ENSDART00000076749
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr5_+_1911814 | 0.18 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr13_+_24280380 | 0.17 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr10_-_26738209 | 0.17 |
ENSDART00000188590
|
fgf13b
|
fibroblast growth factor 13b |
| chr19_-_5669122 | 0.17 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr12_+_48390715 | 0.17 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr22_-_29336268 | 0.16 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr24_+_17260329 | 0.16 |
ENSDART00000129554
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr8_+_36554816 | 0.15 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr20_-_29418620 | 0.15 |
ENSDART00000172634
|
ryr3
|
ryanodine receptor 3 |
| chr21_-_19906388 | 0.15 |
ENSDART00000114969
|
mfhas1
|
malignant fibrous histiocytoma amplified sequence 1 |
| chr5_-_24270989 | 0.15 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr10_+_24643079 | 0.15 |
ENSDART00000139641
ENSDART00000142617 |
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr12_-_25294096 | 0.14 |
ENSDART00000183398
|
hcar1-4
|
hydroxycarboxylic acid receptor 1-4 |
| chr8_-_40276035 | 0.13 |
ENSDART00000185301
ENSDART00000187610 ENSDART00000053158 ENSDART00000162020 |
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr19_+_33093577 | 0.13 |
ENSDART00000180317
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr10_+_26901222 | 0.13 |
ENSDART00000188766
|
ehbp1l1b
|
EH domain binding protein 1-like 1b |
| chr4_+_42175261 | 0.13 |
ENSDART00000162193
|
si:ch211-142b24.2
|
si:ch211-142b24.2 |
| chr4_-_64605318 | 0.12 |
ENSDART00000170391
|
znf1099
|
zinc finger protein 1099 |
| chr17_+_24623561 | 0.12 |
ENSDART00000156734
|
thumpd2
|
THUMP domain containing 2 |
| chr14_+_30774894 | 0.12 |
ENSDART00000023054
|
atl3
|
atlastin 3 |
| chr13_-_12602920 | 0.12 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr18_+_24921587 | 0.12 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr5_+_54585431 | 0.12 |
ENSDART00000171225
|
npr2
|
natriuretic peptide receptor 2 |
| chr25_+_20119466 | 0.12 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr7_-_6357952 | 0.11 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr1_-_35694978 | 0.11 |
ENSDART00000136157
|
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr3_-_21402279 | 0.11 |
ENSDART00000164513
|
CT573446.1
|
|
| chr1_-_43897831 | 0.11 |
ENSDART00000048225
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr9_-_24244383 | 0.10 |
ENSDART00000182407
|
cavin2a
|
caveolae associated protein 2a |
| chr6_-_16561536 | 0.10 |
ENSDART00000167295
|
unc80
|
unc-80 homolog (C. elegans) |
| chr4_+_1530287 | 0.10 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr13_+_8987957 | 0.10 |
ENSDART00000148144
|
hcar1-3
|
hydroxycarboxylic acid receptor 1-3 |
| chr19_-_21716593 | 0.10 |
ENSDART00000155126
|
znf516
|
zinc finger protein 516 |
| chr3_-_4760384 | 0.10 |
ENSDART00000108810
|
CABZ01046997.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of jund_batf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 1.0 | 4.0 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.7 | 2.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.6 | 5.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.6 | 1.7 | GO:0033335 | anal fin development(GO:0033335) |
| 0.5 | 4.4 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.3 | 1.0 | GO:0010935 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.3 | 2.6 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 1.7 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 0.7 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 1.0 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.2 | 0.7 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 1.4 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.7 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.8 | GO:0060420 | regulation of heart growth(GO:0060420) |
| 0.1 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.6 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 1.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 3.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 0.4 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.8 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 0.7 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 4.9 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.4 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.1 | 0.6 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.1 | 0.3 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.1 | 0.6 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.2 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 1.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.0 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 3.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.8 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0036353 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0002544 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 10.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.3 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.2 | GO:0006956 | complement activation(GO:0006956) protein activation cascade(GO:0072376) |
| 0.0 | 2.0 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 8.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 2.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.2 | 4.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.2 | 0.8 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.0 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 1.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.4 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 1.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 4.0 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.2 | 2.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 1.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 5.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 1.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 3.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.4 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.5 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.7 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 4.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 3.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.0 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 1.4 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 3.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 2.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.2 | 4.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.1 | 1.8 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 1.7 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |