Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
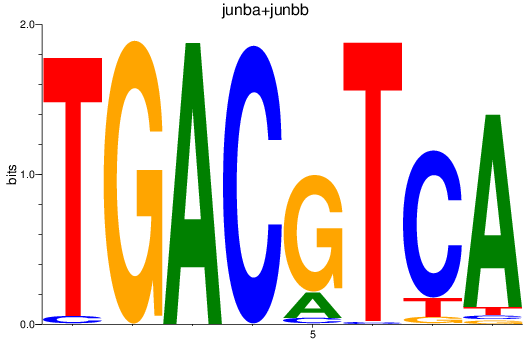
Results for junba+junbb
Z-value: 1.86
Transcription factors associated with junba+junbb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
junba
|
ENSDARG00000074378 | JunB proto-oncogene, AP-1 transcription factor subunit a |
|
junbb
|
ENSDARG00000104773 | JunB proto-oncogene, AP-1 transcription factor subunit b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| junba | dr11_v1_chr1_-_51734524_51734535 | -0.94 | 1.2e-08 | Click! |
| junbb | dr11_v1_chr3_-_7656059_7656059 | -0.88 | 1.9e-06 | Click! |
Activity profile of junba+junbb motif
Sorted Z-values of junba+junbb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_37478418 | 5.05 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr10_-_34915886 | 4.72 |
ENSDART00000141201
ENSDART00000002166 |
ccna1
|
cyclin A1 |
| chr13_+_42309688 | 4.67 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr7_+_46019780 | 4.54 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr25_-_23052707 | 4.48 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr17_+_25187226 | 4.46 |
ENSDART00000148431
|
cln8
|
CLN8, transmembrane ER and ERGIC protein |
| chr12_-_10512911 | 4.28 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr6_+_13232934 | 4.09 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr7_-_26532089 | 4.08 |
ENSDART00000121698
|
senp3b
|
SUMO1/sentrin/SMT3 specific peptidase 3b |
| chr16_-_29387215 | 3.93 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr14_-_41468892 | 3.88 |
ENSDART00000173099
ENSDART00000003170 |
mid1ip1l
|
MID1 interacting protein 1, like |
| chr23_+_19590598 | 3.87 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr23_-_10175898 | 3.79 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr13_+_51710725 | 3.75 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr20_-_36671660 | 3.67 |
ENSDART00000134819
|
slc5a6a
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6a |
| chr8_+_12951155 | 3.64 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr19_-_874888 | 3.62 |
ENSDART00000007206
|
eomesa
|
eomesodermin homolog a |
| chr4_-_20081621 | 3.52 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr22_+_336256 | 3.47 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr19_-_25772980 | 3.44 |
ENSDART00000052393
|
pard6gb
|
par-6 family cell polarity regulator gamma b |
| chr16_+_10557504 | 3.40 |
ENSDART00000091241
|
si:ch73-22o12.1
|
si:ch73-22o12.1 |
| chr3_+_32403758 | 3.38 |
ENSDART00000156982
|
si:ch211-195b15.8
|
si:ch211-195b15.8 |
| chr17_+_32623931 | 3.36 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr1_+_36772348 | 3.33 |
ENSDART00000109314
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr6_+_10333920 | 3.32 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr6_+_27667359 | 3.22 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr24_-_21090447 | 3.22 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr18_-_22094102 | 3.10 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr3_-_15470944 | 3.09 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr11_+_2710530 | 3.08 |
ENSDART00000132768
ENSDART00000030921 ENSDART00000040147 |
mapk14b
|
mitogen-activated protein kinase 14b |
| chr25_-_34845302 | 3.02 |
ENSDART00000039485
|
gabarapl2
|
GABA(A) receptor-associated protein like 2 |
| chr18_+_8917766 | 3.02 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr2_-_37477654 | 2.96 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr4_-_12477224 | 2.94 |
ENSDART00000027756
ENSDART00000182706 ENSDART00000127150 |
arhgef39
|
Rho guanine nucleotide exchange factor (GEF) 39 |
| chr1_+_45707219 | 2.87 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr2_+_9821757 | 2.85 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr22_+_30047245 | 2.84 |
ENSDART00000142857
ENSDART00000141247 ENSDART00000140015 ENSDART00000040538 |
add3a
|
adducin 3 (gamma) a |
| chr6_-_52484566 | 2.84 |
ENSDART00000112146
|
fam83c
|
family with sequence similarity 83, member C |
| chr8_+_12118097 | 2.83 |
ENSDART00000081819
|
endog
|
endonuclease G |
| chr14_-_12106603 | 2.79 |
ENSDART00000054619
|
prps1b
|
phosphoribosyl pyrophosphate synthetase 1B |
| chr7_-_20241346 | 2.78 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr25_+_35891342 | 2.73 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr7_+_57088920 | 2.72 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr3_-_40768548 | 2.71 |
ENSDART00000004923
|
smurf1
|
SMAD specific E3 ubiquitin protein ligase 1 |
| chr25_+_3104959 | 2.70 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr8_-_18613948 | 2.66 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr14_+_30285613 | 2.65 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr23_+_19590006 | 2.63 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr6_+_40992409 | 2.63 |
ENSDART00000151419
|
tgfa
|
transforming growth factor, alpha |
| chr7_-_51749683 | 2.61 |
ENSDART00000083190
|
hdac8
|
histone deacetylase 8 |
| chr11_+_24314148 | 2.59 |
ENSDART00000171491
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr14_+_41318412 | 2.58 |
ENSDART00000064614
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr23_+_12160900 | 2.57 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr13_+_15816573 | 2.53 |
ENSDART00000137061
|
klc1a
|
kinesin light chain 1a |
| chr10_+_4046448 | 2.53 |
ENSDART00000123086
ENSDART00000052268 |
pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr14_+_41318881 | 2.52 |
ENSDART00000192137
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr14_+_41318604 | 2.52 |
ENSDART00000167042
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr1_+_19538299 | 2.51 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr17_-_7792376 | 2.51 |
ENSDART00000064655
|
zbtb2a
|
zinc finger and BTB domain containing 2a |
| chr7_-_69429561 | 2.50 |
ENSDART00000127351
|
atxn1l
|
ataxin 1-like |
| chr19_+_14454306 | 2.49 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr12_+_19199735 | 2.48 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr11_+_24313931 | 2.48 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr20_-_53949798 | 2.46 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr8_+_54274355 | 2.46 |
ENSDART00000067639
|
TMCC1 (1 of many)
|
transmembrane and coiled-coil domain family 1 |
| chr14_-_763744 | 2.45 |
ENSDART00000165856
|
trim35-27
|
tripartite motif containing 35-27 |
| chr6_+_112579 | 2.45 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr17_-_22573311 | 2.45 |
ENSDART00000141523
ENSDART00000140022 ENSDART00000079390 ENSDART00000188644 |
exo1
|
exonuclease 1 |
| chr24_+_25822999 | 2.42 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr7_+_57089354 | 2.42 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr2_-_55779927 | 2.41 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr6_+_49723289 | 2.41 |
ENSDART00000190452
|
stx16
|
syntaxin 16 |
| chr7_-_50764714 | 2.39 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr18_-_30499489 | 2.38 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr21_-_4849029 | 2.37 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr18_+_16749091 | 2.36 |
ENSDART00000061265
|
rnf141
|
ring finger protein 141 |
| chr6_+_3716666 | 2.35 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr20_-_51831657 | 2.34 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr25_+_753364 | 2.34 |
ENSDART00000183804
|
TWF1 (1 of many)
|
twinfilin actin binding protein 1 |
| chr2_+_32743807 | 2.31 |
ENSDART00000022909
|
klhl18
|
kelch-like family member 18 |
| chr8_-_16674584 | 2.28 |
ENSDART00000100727
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr8_+_25959940 | 2.28 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr17_+_24320861 | 2.25 |
ENSDART00000179858
|
otx1
|
orthodenticle homeobox 1 |
| chr5_+_28259655 | 2.24 |
ENSDART00000087684
|
ncaph
|
non-SMC condensin I complex, subunit H |
| chr11_-_43226255 | 2.22 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr20_-_51831816 | 2.20 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr24_-_2423791 | 2.20 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr11_+_506465 | 2.19 |
ENSDART00000082519
|
raf1b
|
Raf-1 proto-oncogene, serine/threonine kinase b |
| chr25_+_17860798 | 2.18 |
ENSDART00000146845
|
pth1a
|
parathyroid hormone 1a |
| chr3_-_61494840 | 2.18 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr17_+_25849332 | 2.16 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr2_+_24762567 | 2.15 |
ENSDART00000078866
|
ifi30
|
interferon, gamma-inducible protein 30 |
| chr12_+_14079097 | 2.14 |
ENSDART00000078033
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr16_+_43077909 | 2.09 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr4_+_9011825 | 2.09 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr2_-_37352514 | 2.09 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr5_+_26204561 | 2.08 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr1_+_49878000 | 2.08 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr12_+_38807604 | 2.05 |
ENSDART00000155563
|
abca5
|
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr14_-_33478963 | 2.05 |
ENSDART00000132813
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr3_-_21137362 | 2.04 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr7_+_10562118 | 2.02 |
ENSDART00000185188
ENSDART00000168801 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr17_+_24064014 | 2.01 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr17_+_50701748 | 2.00 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr21_+_18907102 | 2.00 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr19_-_12965020 | 2.00 |
ENSDART00000128975
|
slc25a32a
|
solute carrier family 25 (mitochondrial folate carrier), member 32a |
| chr13_+_22719789 | 1.99 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr23_-_29357764 | 1.97 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr4_+_9011448 | 1.96 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr3_-_25369557 | 1.96 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr13_+_28785814 | 1.96 |
ENSDART00000039028
|
nsmce4a
|
NSE4 homolog A, SMC5-SMC6 complex component |
| chr1_-_59240975 | 1.96 |
ENSDART00000166170
|
mvb12a
|
multivesicular body subunit 12A |
| chr10_-_28380919 | 1.94 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr1_+_49266886 | 1.93 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr9_+_50110763 | 1.93 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr3_+_33745014 | 1.93 |
ENSDART00000159966
|
nacc1a
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing a |
| chr10_+_5689510 | 1.93 |
ENSDART00000183217
ENSDART00000172632 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr4_-_16628801 | 1.92 |
ENSDART00000040708
ENSDART00000064009 |
caprin2
|
caprin family member 2 |
| chr8_+_50190742 | 1.91 |
ENSDART00000099863
|
slc25a37
|
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr11_+_2687395 | 1.91 |
ENSDART00000082510
|
b3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr5_+_33287611 | 1.91 |
ENSDART00000125093
ENSDART00000146759 |
med22
|
mediator complex subunit 22 |
| chr23_-_24542952 | 1.91 |
ENSDART00000088777
|
atp13a2
|
ATPase 13A2 |
| chr2_-_45153531 | 1.91 |
ENSDART00000109890
|
capn10
|
calpain 10 |
| chr11_+_31380495 | 1.90 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr5_-_13315726 | 1.90 |
ENSDART00000143364
|
sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_-_49504023 | 1.90 |
ENSDART00000168108
|
prkacaa
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate a |
| chr23_-_17509656 | 1.90 |
ENSDART00000148423
|
dnajc5ab
|
DnaJ (Hsp40) homolog, subfamily C, member 5ab |
| chr5_+_20366453 | 1.89 |
ENSDART00000193141
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr7_+_34549198 | 1.89 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr5_-_29531948 | 1.89 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr24_+_37338169 | 1.87 |
ENSDART00000141771
|
gnptg
|
N-acetylglucosamine-1-phosphate transferase, gamma subunit |
| chr16_-_42066523 | 1.85 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr11_-_27827442 | 1.83 |
ENSDART00000121847
ENSDART00000132018 ENSDART00000145744 ENSDART00000134677 ENSDART00000130800 |
cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1 |
| chr3_-_43650189 | 1.83 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr5_-_25576462 | 1.83 |
ENSDART00000165147
|
si:dkey-229d2.4
|
si:dkey-229d2.4 |
| chr23_+_24973773 | 1.83 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr15_-_41689981 | 1.83 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr22_-_10539180 | 1.83 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr24_+_14801844 | 1.82 |
ENSDART00000141620
|
pi15a
|
peptidase inhibitor 15a |
| chr15_-_8309207 | 1.82 |
ENSDART00000143880
ENSDART00000061351 |
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr25_+_17860962 | 1.80 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr1_+_52792439 | 1.78 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr5_+_33289057 | 1.77 |
ENSDART00000123210
|
med22
|
mediator complex subunit 22 |
| chr3_+_19245804 | 1.77 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr11_-_44647286 | 1.76 |
ENSDART00000169329
ENSDART00000158939 |
tomm20b
|
translocase of outer mitochondrial membrane 20b |
| chr17_+_17764979 | 1.76 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr15_+_34963316 | 1.76 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr16_-_17347727 | 1.75 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr8_-_13678415 | 1.73 |
ENSDART00000134153
ENSDART00000143331 |
si:dkey-258f14.3
|
si:dkey-258f14.3 |
| chr4_-_25515154 | 1.71 |
ENSDART00000186524
|
rbm17
|
RNA binding motif protein 17 |
| chr18_+_17493859 | 1.71 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr19_-_6840506 | 1.70 |
ENSDART00000081568
|
tcf19l
|
transcription factor 19 (SC1), like |
| chr8_+_47683539 | 1.70 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr6_-_1587291 | 1.70 |
ENSDART00000067592
ENSDART00000178877 |
zgc:123305
|
zgc:123305 |
| chr25_-_36263115 | 1.70 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr13_+_534453 | 1.70 |
ENSDART00000147909
|
wu:fc17b08
|
wu:fc17b08 |
| chr9_-_30259447 | 1.68 |
ENSDART00000140585
ENSDART00000145052 ENSDART00000129926 |
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr4_-_25515513 | 1.68 |
ENSDART00000142276
ENSDART00000044043 |
rbm17
|
RNA binding motif protein 17 |
| chr16_-_52540056 | 1.68 |
ENSDART00000188304
|
CR293507.1
|
|
| chr7_+_34549377 | 1.68 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr16_+_27614989 | 1.68 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr21_+_22558187 | 1.67 |
ENSDART00000167599
|
chek1
|
checkpoint kinase 1 |
| chr9_-_30259295 | 1.67 |
ENSDART00000139106
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr8_+_49433663 | 1.67 |
ENSDART00000140481
ENSDART00000180714 |
nfu1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr16_-_41535690 | 1.65 |
ENSDART00000102662
|
rpp25l
|
ribonuclease P/MRP 25 subunit-like |
| chr13_-_17943135 | 1.64 |
ENSDART00000176027
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr5_-_20921677 | 1.64 |
ENSDART00000158030
|
si:ch211-225b11.4
|
si:ch211-225b11.4 |
| chr3_+_40576447 | 1.62 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr17_+_30591287 | 1.62 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr24_-_40901410 | 1.61 |
ENSDART00000170688
|
wdr48a
|
WD repeat domain 48a |
| chr21_-_30166097 | 1.61 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr19_-_31707892 | 1.61 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr8_-_13735572 | 1.61 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr16_+_46410520 | 1.60 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr13_-_23956178 | 1.60 |
ENSDART00000133646
|
phactr2
|
phosphatase and actin regulator 2 |
| chr2_+_38055529 | 1.60 |
ENSDART00000145642
|
si:rp71-1g18.1
|
si:rp71-1g18.1 |
| chr10_+_7593185 | 1.59 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr21_-_43398457 | 1.59 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr13_+_46941930 | 1.58 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr9_+_33334501 | 1.57 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr7_-_51749895 | 1.57 |
ENSDART00000175523
ENSDART00000189639 |
hdac8
|
histone deacetylase 8 |
| chr23_-_10745288 | 1.57 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr7_+_38808027 | 1.56 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr2_-_19354622 | 1.56 |
ENSDART00000168627
|
zfyve9a
|
zinc finger, FYVE domain containing 9a |
| chr13_+_29925397 | 1.56 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr21_-_43398122 | 1.56 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr24_-_37338162 | 1.55 |
ENSDART00000056303
|
tsr3
|
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr11_-_44898129 | 1.52 |
ENSDART00000157615
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr9_-_6502491 | 1.52 |
ENSDART00000102672
|
nck2a
|
NCK adaptor protein 2a |
| chr18_-_8380090 | 1.52 |
ENSDART00000141581
ENSDART00000081143 |
sephs1
|
selenophosphate synthetase 1 |
| chr13_-_23956361 | 1.52 |
ENSDART00000101150
|
phactr2
|
phosphatase and actin regulator 2 |
| chr16_-_12060770 | 1.52 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr25_-_37084032 | 1.51 |
ENSDART00000025494
|
hprt1l
|
hypoxanthine phosphoribosyltransferase 1, like |
| chr6_+_33537267 | 1.51 |
ENSDART00000040334
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr12_+_33320504 | 1.50 |
ENSDART00000021491
|
csnk1db
|
casein kinase 1, delta b |
| chr13_-_23612324 | 1.50 |
ENSDART00000136406
ENSDART00000005004 |
prim2
|
DNA primase subunit 2 |
| chr16_-_41667101 | 1.50 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr10_+_1681518 | 1.50 |
ENSDART00000018532
|
triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr15_-_41689684 | 1.50 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr20_+_9223514 | 1.49 |
ENSDART00000023293
|
kcnk5b
|
potassium channel, subfamily K, member 5b |
Network of associatons between targets according to the STRING database.
First level regulatory network of junba+junbb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 1.2 | 3.6 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 1.0 | 5.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.9 | 3.7 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.9 | 5.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.8 | 2.4 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.7 | 2.8 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.7 | 4.7 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.7 | 9.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.7 | 2.0 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.6 | 1.9 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.6 | 5.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.6 | 1.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.5 | 1.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.5 | 4.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.5 | 2.0 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.5 | 1.5 | GO:0046099 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.5 | 1.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.5 | 1.8 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.4 | 2.1 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.4 | 1.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.4 | 5.3 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.4 | 2.8 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.4 | 1.6 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.4 | 1.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.4 | 1.5 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.4 | 1.8 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.4 | 0.4 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.4 | 2.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.4 | 1.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.4 | 1.8 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.4 | 1.8 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.3 | 2.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.3 | 2.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 3.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.3 | 1.6 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 | 1.9 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.3 | 1.6 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.3 | 3.4 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.3 | 3.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 1.2 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.3 | 0.9 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.3 | 1.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.3 | 3.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.3 | 1.7 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 0.8 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.3 | 2.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.3 | 1.9 | GO:0071333 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.3 | 1.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 1.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.3 | 1.0 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.3 | 2.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 1.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 12.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.3 | 4.5 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.3 | 1.0 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 1.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 1.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 4.0 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.2 | 0.9 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 1.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 2.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.2 | 1.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 2.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.6 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.2 | 1.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 0.8 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 1.3 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 2.0 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 0.6 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 2.9 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.2 | 2.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.2 | 2.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 2.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 2.6 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 | 1.2 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 1.7 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 2.7 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.2 | 1.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.2 | 2.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.2 | 0.6 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.2 | 1.2 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.2 | 3.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.2 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.4 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.2 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.0 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 2.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.7 | GO:0033152 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 1.0 | GO:2001270 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 3.8 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 2.4 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.8 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 2.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.8 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 3.0 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 2.6 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.5 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 4.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 1.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 4.3 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.1 | 0.7 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.1 | 0.4 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 3.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 2.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 2.5 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.1 | 0.5 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.8 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.1 | 0.8 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 3.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 2.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 2.0 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 2.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 6.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.1 | 4.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.1 | 0.8 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 5.1 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.1 | 2.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 2.1 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.6 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.5 | GO:0019859 | thymine catabolic process(GO:0006210) valine catabolic process(GO:0006574) thymine metabolic process(GO:0019859) |
| 0.1 | 0.8 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.7 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 3.0 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.6 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 1.9 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.1 | GO:2000369 | positive regulation of Notch signaling pathway(GO:0045747) regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 3.2 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 2.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.5 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 2.0 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 0.3 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.4 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 1.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 2.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 1.9 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.1 | 1.2 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 1.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 2.2 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.1 | 1.8 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.1 | 5.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 1.2 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.1 | 1.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.5 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.7 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 6.4 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 0.3 | GO:0045932 | negative regulation of muscle contraction(GO:0045932) |
| 0.1 | 0.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 1.9 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.1 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 3.9 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.9 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 2.3 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.2 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.3 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 2.0 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 1.9 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0072028 | nephron morphogenesis(GO:0072028) |
| 0.0 | 0.1 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 3.1 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.5 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 2.6 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.5 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 1.1 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 3.8 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 0.7 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 1.0 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 2.7 | GO:0006417 | regulation of translation(GO:0006417) |
| 0.0 | 3.0 | GO:0030335 | positive regulation of cell migration(GO:0030335) |
| 0.0 | 2.4 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.0 | 1.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 1.0 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.8 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 1.0 | GO:0050684 | regulation of mRNA processing(GO:0050684) regulation of mRNA metabolic process(GO:1903311) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.5 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.4 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.7 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 1.4 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 2.4 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.7 | GO:0006661 | phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 | 0.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.0 | GO:0022406 | membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.4 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 1.1 | 3.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 1.1 | 4.5 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.8 | 4.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.7 | 2.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.7 | 2.7 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 4.1 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.5 | 1.5 | GO:1990077 | primosome complex(GO:1990077) |
| 0.4 | 2.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 2.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.4 | 1.6 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 3.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 2.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 2.6 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.2 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 1.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.3 | 2.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 1.6 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 1.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 1.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.7 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.2 | 4.3 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.2 | 1.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.2 | 1.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.2 | 0.8 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.2 | 1.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 3.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 3.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.2 | 4.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.2 | 2.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 1.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.2 | 4.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.5 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.2 | 1.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) outer mitochondrial membrane protein complex(GO:0098799) |
| 0.2 | 0.8 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.2 | 2.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 2.2 | GO:0071010 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 4.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.0 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 1.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 3.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 2.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 2.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.8 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.7 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.8 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0035032 | extrinsic component of vacuolar membrane(GO:0000306) phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 8.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 4.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 2.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 3.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 5.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 1.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 3.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.4 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 4.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 4.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 2.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 4.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 3.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 4.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 28.3 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 2.6 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 3.7 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 2.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 6.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 6.9 | GO:0005740 | mitochondrial envelope(GO:0005740) |
| 0.0 | 5.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.5 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.0 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.9 | 3.7 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.8 | 2.4 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.7 | 3.6 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.7 | 2.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.7 | 2.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.7 | 9.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.6 | 2.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.5 | 2.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.5 | 1.5 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.5 | 1.9 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.5 | 2.8 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.5 | 2.3 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.5 | 1.4 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.4 | 4.0 | GO:0097001 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.4 | 1.8 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.4 | 5.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.4 | 1.9 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.4 | 1.9 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.4 | 1.5 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.4 | 6.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 2.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 2.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.4 | 4.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.3 | 1.3 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.3 | 1.9 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.3 | 2.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 0.9 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.3 | 1.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 1.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 5.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 1.0 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.2 | 1.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.7 | GO:1901611 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.2 | 1.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.2 | 5.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 13.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.0 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 1.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 0.8 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 1.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.2 | 5.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 4.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.2 | 0.7 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.2 | 1.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 2.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 3.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 1.0 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.2 | 2.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.2 | 0.8 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.2 | 2.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 2.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 0.8 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.2 | 2.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.2 | 0.5 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.2 | 1.4 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 3.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.3 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.3 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 2.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.9 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 1.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 1.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 1.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 5.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 1.8 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 3.5 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 4.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.9 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 2.2 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 1.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 2.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 2.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.1 | 4.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.1 | 0.6 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 2.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.6 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.5 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.1 | 2.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 4.4 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 2.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.3 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 1.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 1.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.8 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 3.2 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 1.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 1.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 1.3 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.3 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 10.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 2.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.4 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.9 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 1.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 9.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 2.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 3.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 1.3 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 5.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 2.5 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.5 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 1.3 | GO:0004518 | nuclease activity(GO:0004518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 9.8 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.3 | 3.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 1.8 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 13.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.2 | 1.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 3.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 1.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 5.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 2.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 3.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 4.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.1 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 0.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 4.7 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 2.1 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 2.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 1.0 | 9.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 2.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 10.8 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.3 | 4.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.3 | 4.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.2 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 2.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 2.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 2.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 4.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 1.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 2.0 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.2 | 2.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 2.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 1.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 5.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 4.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 2.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 0.7 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 3.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 3.2 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.1 | 1.4 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.1 | 2.9 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 2.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 0.6 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 4.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.2 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.1 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.3 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 1.8 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.3 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.8 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |