Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
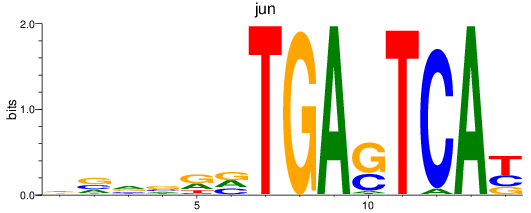
Results for jun
Z-value: 0.40
Transcription factors associated with jun
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jun
|
ENSDARG00000043531 | Jun proto-oncogene, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jun | dr11_v1_chr20_+_15552657_15552657 | 0.86 | 5.2e-06 | Click! |
Activity profile of jun motif
Sorted Z-values of jun motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_24488403 | 1.34 |
ENSDART00000052421
|
txnipa
|
thioredoxin interacting protein a |
| chr11_-_24681292 | 1.32 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr20_+_26880668 | 1.25 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr3_-_61593274 | 1.23 |
ENSDART00000154132
ENSDART00000055071 |
nptx2a
|
neuronal pentraxin 2a |
| chr24_+_31361407 | 1.12 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr25_-_1126311 | 1.04 |
ENSDART00000161863
|
spg11
|
spastic paraplegia 11 |
| chr7_-_52963493 | 1.02 |
ENSDART00000052029
|
cart3
|
cocaine- and amphetamine-regulated transcript 3 |
| chr21_+_22845317 | 1.02 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr6_-_40842768 | 0.99 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr22_-_10110959 | 0.87 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr10_+_31222433 | 0.83 |
ENSDART00000185080
|
tmem218
|
transmembrane protein 218 |
| chr10_+_31222656 | 0.83 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr13_+_6086730 | 0.82 |
ENSDART00000049328
|
fam120b
|
family with sequence similarity 120B |
| chr22_+_16497670 | 0.80 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr8_+_17933475 | 0.79 |
ENSDART00000100651
|
erich3
|
glutamate-rich 3 |
| chr1_+_44340124 | 0.77 |
ENSDART00000160696
|
CT033803.2
|
|
| chr15_-_46779934 | 0.76 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr3_+_39922602 | 0.74 |
ENSDART00000193937
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr16_+_42829735 | 0.73 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr9_-_710896 | 0.71 |
ENSDART00000180478
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr1_-_59252973 | 0.71 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr1_+_44360973 | 0.69 |
ENSDART00000167568
|
si:ch211-165a10.5
|
si:ch211-165a10.5 |
| chr6_+_48348415 | 0.68 |
ENSDART00000064826
|
mov10a
|
Mov10 RISC complex RNA helicase a |
| chr4_-_6809323 | 0.68 |
ENSDART00000099467
|
ifrd1
|
interferon-related developmental regulator 1 |
| chr2_+_17524278 | 0.67 |
ENSDART00000165633
|
pimr196
|
Pim proto-oncogene, serine/threonine kinase, related 196 |
| chr7_+_20587506 | 0.66 |
ENSDART00000172416
ENSDART00000170633 |
si:dkey-19b23.7
|
si:dkey-19b23.7 |
| chr1_-_40123943 | 0.64 |
ENSDART00000146917
|
si:ch211-113e8.10
|
si:ch211-113e8.10 |
| chr5_-_57480660 | 0.60 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr13_-_24328794 | 0.58 |
ENSDART00000145351
|
si:ch211-202m22.1
|
si:ch211-202m22.1 |
| chr5_-_69523816 | 0.56 |
ENSDART00000112692
|
si:ch211-157p22.10
|
si:ch211-157p22.10 |
| chr1_+_44260085 | 0.56 |
ENSDART00000171416
|
si:dkey-59p5.1
|
si:dkey-59p5.1 |
| chr16_+_41517188 | 0.55 |
ENSDART00000049976
|
si:dkey-11p23.7
|
si:dkey-11p23.7 |
| chr18_+_20838786 | 0.54 |
ENSDART00000138692
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr1_+_44249616 | 0.53 |
ENSDART00000164173
|
si:ch211-165a10.10
|
si:ch211-165a10.10 |
| chr12_+_1455147 | 0.53 |
ENSDART00000018752
|
cops3
|
COP9 signalosome subunit 3 |
| chr24_+_24923166 | 0.53 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr25_-_7753207 | 0.51 |
ENSDART00000126499
|
phf21ab
|
PHD finger protein 21Ab |
| chr12_+_20506197 | 0.51 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr5_+_37087583 | 0.49 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr21_-_30545121 | 0.49 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr10_-_27566481 | 0.49 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr16_-_45225520 | 0.48 |
ENSDART00000158855
|
fxyd1
|
FXYD domain containing ion transport regulator 1 (phospholemman) |
| chr6_+_33885828 | 0.46 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr12_+_7497882 | 0.46 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr1_+_2190714 | 0.45 |
ENSDART00000132126
|
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr8_+_50534948 | 0.45 |
ENSDART00000174435
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr16_+_10972733 | 0.44 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr24_-_33693070 | 0.44 |
ENSDART00000137483
|
si:dkeyp-87a6.2
|
si:dkeyp-87a6.2 |
| chr16_-_21785261 | 0.44 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr23_+_24600494 | 0.44 |
ENSDART00000184854
|
nckap5l
|
NCK-associated protein 5-like |
| chr7_+_5566582 | 0.43 |
ENSDART00000145458
|
si:ch211-165f21.4
|
si:ch211-165f21.4 |
| chr3_+_7617353 | 0.42 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr11_+_2416064 | 0.42 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr14_+_33882973 | 0.41 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr11_+_13630107 | 0.40 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr10_+_31344227 | 0.40 |
ENSDART00000184470
|
AL845320.2
|
|
| chr1_+_6640437 | 0.40 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr4_-_16412084 | 0.40 |
ENSDART00000188460
|
dcn
|
decorin |
| chr4_+_1600034 | 0.40 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr7_+_14005111 | 0.39 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr14_+_1240235 | 0.39 |
ENSDART00000127477
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr4_+_357810 | 0.39 |
ENSDART00000163436
ENSDART00000103645 |
tmem181
|
transmembrane protein 181 |
| chr13_-_37649595 | 0.39 |
ENSDART00000115354
|
si:dkey-188i13.10
|
si:dkey-188i13.10 |
| chr17_+_10817675 | 0.39 |
ENSDART00000156090
|
pimr177
|
Pim proto-oncogene, serine/threonine kinase, related 177 |
| chr3_+_56905103 | 0.39 |
ENSDART00000124728
ENSDART00000154408 |
otop2
|
otopetrin 2 |
| chr2_-_293793 | 0.39 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr8_-_23573084 | 0.38 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr7_-_35432901 | 0.38 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr1_+_44350628 | 0.38 |
ENSDART00000171939
|
CT033803.1
|
|
| chr14_+_1240604 | 0.37 |
ENSDART00000188258
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr23_-_4915118 | 0.37 |
ENSDART00000060714
|
atp6ap1a
|
ATPase H+ transporting accessory protein 1a |
| chr22_+_24220937 | 0.37 |
ENSDART00000165380
|
uchl5
|
ubiquitin carboxyl-terminal hydrolase L5 |
| chr5_+_15819651 | 0.37 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr4_-_57572054 | 0.37 |
ENSDART00000158610
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr9_-_2572790 | 0.37 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr17_+_7524389 | 0.37 |
ENSDART00000067579
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr7_+_22688781 | 0.37 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr13_+_10621257 | 0.36 |
ENSDART00000008603
|
prepl
|
prolyl endopeptidase-like |
| chr20_-_25626198 | 0.36 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr13_+_9213805 | 0.36 |
ENSDART00000131455
|
pimr158
|
Pim proto-oncogene, serine/threonine kinase, related 158 |
| chr16_+_42830152 | 0.36 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr14_-_26436760 | 0.35 |
ENSDART00000088677
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr16_-_13818061 | 0.35 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr3_+_24986145 | 0.35 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr4_-_9637398 | 0.35 |
ENSDART00000150253
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr20_-_40487208 | 0.35 |
ENSDART00000075070
ENSDART00000142029 |
hsf2
|
heat shock transcription factor 2 |
| chr23_+_16814968 | 0.35 |
ENSDART00000161299
|
zgc:100832
|
zgc:100832 |
| chr19_-_41404870 | 0.35 |
ENSDART00000167772
|
sem1
|
SEM1, 26S proteasome complex subunit |
| chr24_+_38522254 | 0.35 |
ENSDART00000156189
|
si:ch1073-66l23.1
|
si:ch1073-66l23.1 |
| chr2_+_48288461 | 0.33 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr17_-_6536305 | 0.33 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr22_+_36547670 | 0.33 |
ENSDART00000125991
ENSDART00000186372 |
armc9
|
armadillo repeat containing 9 |
| chr10_+_26747755 | 0.32 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr5_-_19052184 | 0.32 |
ENSDART00000133330
|
fam214b
|
family with sequence similarity 214, member B |
| chr23_+_28128453 | 0.32 |
ENSDART00000182618
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr8_+_50531709 | 0.32 |
ENSDART00000193352
|
PEBP4
|
phosphatidylethanolamine binding protein 4 |
| chr16_+_49005321 | 0.32 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr14_+_49382180 | 0.32 |
ENSDART00000158329
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr14_-_26436951 | 0.32 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr15_-_28223757 | 0.32 |
ENSDART00000110969
ENSDART00000138401 |
scarf1
|
scavenger receptor class F, member 1 |
| chr19_+_23932259 | 0.31 |
ENSDART00000139040
|
si:dkey-222b8.1
|
si:dkey-222b8.1 |
| chr8_+_20037368 | 0.30 |
ENSDART00000029175
ENSDART00000170792 |
fcho1
|
FCH domain only 1 |
| chr17_+_12255642 | 0.30 |
ENSDART00000155175
|
pimr175
|
Pim proto-oncogene, serine/threonine kinase, related 175 |
| chr17_+_51682429 | 0.30 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr23_-_37113396 | 0.30 |
ENSDART00000102886
ENSDART00000134461 |
zgc:193690
|
zgc:193690 |
| chr8_+_40210398 | 0.30 |
ENSDART00000167612
|
rnf34a
|
ring finger protein 34a |
| chr14_+_46216703 | 0.29 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr25_-_35045250 | 0.29 |
ENSDART00000156508
ENSDART00000126590 |
zgc:114046
|
zgc:114046 |
| chr11_-_22371105 | 0.29 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr1_+_52518176 | 0.29 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr15_-_35134918 | 0.29 |
ENSDART00000157196
|
si:ch211-272b8.7
|
si:ch211-272b8.7 |
| chr16_+_54674556 | 0.29 |
ENSDART00000167040
|
pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr6_+_52350443 | 0.29 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr23_+_35714574 | 0.28 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr8_-_18667693 | 0.28 |
ENSDART00000100516
|
stap2b
|
signal transducing adaptor family member 2b |
| chr11_-_25324534 | 0.28 |
ENSDART00000158598
|
si:ch211-232b12.5
|
si:ch211-232b12.5 |
| chr3_-_26017592 | 0.27 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr15_+_9327252 | 0.27 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr16_-_48673938 | 0.27 |
ENSDART00000156969
|
notchl
|
notch homolog, like |
| chr5_-_34616599 | 0.27 |
ENSDART00000050271
ENSDART00000097975 |
hexb
|
hexosaminidase B (beta polypeptide) |
| chr9_+_26199674 | 0.27 |
ENSDART00000139510
|
si:dkey-111i23.1
|
si:dkey-111i23.1 |
| chr1_+_44239558 | 0.27 |
ENSDART00000100335
|
si:dkey-59p5.2
|
si:dkey-59p5.2 |
| chr5_+_23599259 | 0.27 |
ENSDART00000138902
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr20_+_51478939 | 0.27 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr5_-_26879302 | 0.27 |
ENSDART00000098571
ENSDART00000139086 |
zgc:64051
|
zgc:64051 |
| chr21_+_8341774 | 0.27 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr5_-_21888368 | 0.27 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr19_-_7495006 | 0.27 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr21_-_21020708 | 0.26 |
ENSDART00000064032
|
eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chr1_+_41849152 | 0.26 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr4_-_20208166 | 0.26 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr20_+_25625872 | 0.26 |
ENSDART00000078385
|
ppat
|
phosphoribosyl pyrophosphate amidotransferase |
| chr5_-_55623443 | 0.26 |
ENSDART00000005671
ENSDART00000176341 |
hnrpkl
|
heterogeneous nuclear ribonucleoprotein K, like |
| chr15_+_714203 | 0.26 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr7_+_33172066 | 0.26 |
ENSDART00000174013
|
si:ch211-194p6.12
|
si:ch211-194p6.12 |
| chr1_-_25679339 | 0.26 |
ENSDART00000161703
ENSDART00000054230 |
fgg
|
fibrinogen gamma chain |
| chr2_+_47581997 | 0.26 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr9_-_2573121 | 0.26 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr10_-_34741738 | 0.26 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr14_+_6287871 | 0.25 |
ENSDART00000105344
|
tbc1d2
|
TBC1 domain family, member 2 |
| chr19_+_22216778 | 0.25 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr10_+_28428222 | 0.25 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr3_-_58189429 | 0.25 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr21_+_33459524 | 0.25 |
ENSDART00000053205
|
cd74b
|
CD74 molecule, major histocompatibility complex, class II invariant chain b |
| chr5_-_22501663 | 0.25 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr4_-_17822447 | 0.25 |
ENSDART00000185502
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr4_+_77971104 | 0.24 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr8_+_3530761 | 0.24 |
ENSDART00000081272
|
gcn1
|
GCN1 eIF2 alpha kinase activator homolog |
| chr21_+_17880511 | 0.24 |
ENSDART00000080481
|
rxraa
|
retinoid X receptor, alpha a |
| chr10_+_38610741 | 0.24 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr10_+_187760 | 0.24 |
ENSDART00000161007
ENSDART00000160651 |
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr25_+_10485103 | 0.24 |
ENSDART00000104576
|
psmd13
|
proteasome 26S subunit, non-ATPase 13 |
| chr17_-_43399896 | 0.23 |
ENSDART00000156033
ENSDART00000156418 |
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr8_-_25338709 | 0.23 |
ENSDART00000131616
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr13_+_50368668 | 0.23 |
ENSDART00000127610
|
dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr24_-_25186895 | 0.23 |
ENSDART00000081029
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr3_-_26017831 | 0.23 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr13_-_17723417 | 0.23 |
ENSDART00000183834
|
vdac2
|
voltage-dependent anion channel 2 |
| chr4_-_69189894 | 0.22 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr8_+_41038141 | 0.22 |
ENSDART00000075620
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr19_+_10396042 | 0.22 |
ENSDART00000028048
ENSDART00000151735 |
necap1
|
NECAP endocytosis associated 1 |
| chr7_-_28696556 | 0.22 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr3_+_27027781 | 0.22 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr13_+_9237493 | 0.22 |
ENSDART00000140240
|
pimr159
|
Pim proto-oncogene, serine/threonine kinase, related 159 |
| chr7_-_22941472 | 0.22 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr9_-_2594410 | 0.22 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr23_-_37113215 | 0.22 |
ENSDART00000146835
|
zgc:193690
|
zgc:193690 |
| chr3_+_26064091 | 0.21 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr10_+_40583930 | 0.21 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr15_-_30832171 | 0.21 |
ENSDART00000152414
ENSDART00000152790 |
msi2b
|
musashi RNA-binding protein 2b |
| chr6_-_60079551 | 0.21 |
ENSDART00000154753
|
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr2_-_49997055 | 0.21 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr3_+_32118670 | 0.21 |
ENSDART00000055287
ENSDART00000111688 |
zgc:109934
|
zgc:109934 |
| chr3_-_39648021 | 0.20 |
ENSDART00000055171
|
grapa
|
GRB2-related adaptor protein a |
| chr11_-_10770053 | 0.20 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr5_+_37785152 | 0.20 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr20_-_26531850 | 0.20 |
ENSDART00000183317
ENSDART00000131994 |
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr2_-_898899 | 0.20 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr4_+_16715267 | 0.20 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr18_+_3052408 | 0.20 |
ENSDART00000181563
|
kctd14
|
potassium channel tetramerization domain containing 14 |
| chr17_-_7861219 | 0.20 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr20_+_51479263 | 0.19 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr5_+_15350954 | 0.19 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr6_-_49063085 | 0.19 |
ENSDART00000156124
|
si:ch211-105j21.9
|
si:ch211-105j21.9 |
| chr23_+_42336084 | 0.19 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr2_+_30379650 | 0.19 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr23_+_4324625 | 0.19 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr25_+_2282265 | 0.19 |
ENSDART00000076417
|
cdkn1bb
|
cyclin-dependent kinase inhibitor 1Bb |
| chr12_+_29173523 | 0.19 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr2_-_3038904 | 0.19 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr5_+_45677781 | 0.19 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr2_-_37837472 | 0.18 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr7_+_34492744 | 0.18 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr20_+_7084154 | 0.18 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr8_-_31606514 | 0.18 |
ENSDART00000018886
|
ghra
|
growth hormone receptor a |
| chr16_+_50006872 | 0.17 |
ENSDART00000157100
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr20_-_26532167 | 0.17 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr4_+_47656992 | 0.17 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr19_-_27564458 | 0.16 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr24_-_2736809 | 0.16 |
ENSDART00000171805
|
fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
Network of associatons between targets according to the STRING database.
First level regulatory network of jun
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.2 | 1.0 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 0.5 | GO:0043576 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 1.0 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.1 | 0.5 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.5 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.9 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:1902230 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.2 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.8 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.2 | GO:0010991 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.2 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.8 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.4 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 1.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.3 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.7 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0060959 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.4 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.2 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.4 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 5.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.1 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.4 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.0 | GO:0099540 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:1902882 | regulation of response to oxidative stress(GO:1902882) |
| 0.0 | 0.5 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 1.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.3 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.0 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.3 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 0.3 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.1 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.4 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.6 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 1.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.4 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.7 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.6 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.3 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 1.0 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.2 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.3 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.4 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.2 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.4 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.8 | GO:0008308 | voltage-gated chloride channel activity(GO:0005247) voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.0 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 1.0 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |