Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
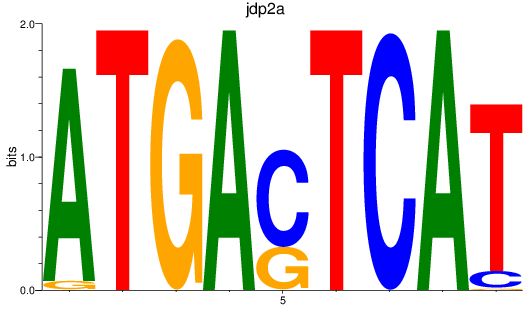
Results for jdp2a
Z-value: 0.45
Transcription factors associated with jdp2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
jdp2a
|
ENSDARG00000040137 | Jun dimerization protein 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| jdp2a | dr11_v1_chr17_-_50220228_50220228 | -0.03 | 9.0e-01 | Click! |
Activity profile of jdp2a motif
Sorted Z-values of jdp2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_34402839 | 1.75 |
ENSDART00000180173
ENSDART00000191588 ENSDART00000186376 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr25_+_30196039 | 1.21 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr5_+_34549845 | 1.19 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr15_+_25489406 | 1.18 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr21_+_13233377 | 1.10 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr7_+_44802353 | 1.09 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr9_+_33154841 | 1.07 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr2_-_23391266 | 1.04 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr2_-_32738535 | 1.02 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr20_+_28364742 | 0.93 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr7_-_60351876 | 0.86 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr11_-_39118882 | 0.85 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr11_+_19370717 | 0.85 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr7_-_60351537 | 0.84 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr12_-_35386910 | 0.83 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr21_+_37477001 | 0.83 |
ENSDART00000114778
|
amot
|
angiomotin |
| chr6_-_33685325 | 0.82 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr2_+_15100742 | 0.81 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr16_+_23403602 | 0.77 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr22_-_23267893 | 0.77 |
ENSDART00000105613
|
si:dkey-121a9.3
|
si:dkey-121a9.3 |
| chr6_+_59832786 | 0.74 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr6_-_37744430 | 0.73 |
ENSDART00000150177
ENSDART00000149722 |
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr21_+_25765734 | 0.72 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr1_-_55116453 | 0.72 |
ENSDART00000142348
|
sertad2a
|
SERTA domain containing 2a |
| chr5_-_54482245 | 0.71 |
ENSDART00000177470
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr13_+_7575563 | 0.71 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr6_+_44197348 | 0.69 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr9_+_33334501 | 0.69 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr6_+_44197099 | 0.66 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr16_-_24832038 | 0.66 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr8_-_22288004 | 0.65 |
ENSDART00000100042
|
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr8_-_22288258 | 0.65 |
ENSDART00000140978
ENSDART00000100046 |
si:ch211-147a11.3
|
si:ch211-147a11.3 |
| chr15_+_17343319 | 0.65 |
ENSDART00000018461
|
vmp1
|
vacuole membrane protein 1 |
| chr3_+_35498119 | 0.64 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr24_-_38192003 | 0.62 |
ENSDART00000109975
|
crp7
|
C-reactive protein 7 |
| chr10_+_5954787 | 0.62 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr10_-_32877348 | 0.62 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr15_+_29709382 | 0.62 |
ENSDART00000099956
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr9_-_22057658 | 0.61 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr2_-_51087077 | 0.59 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr18_+_16750080 | 0.58 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr19_+_26072624 | 0.58 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr5_+_57480014 | 0.57 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr24_-_21973365 | 0.57 |
ENSDART00000081204
ENSDART00000030592 |
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr2_+_9821757 | 0.57 |
ENSDART00000018408
ENSDART00000141227 ENSDART00000144681 ENSDART00000148227 |
anxa13l
|
annexin A13, like |
| chr20_-_16548912 | 0.56 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr19_-_31035155 | 0.56 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr1_+_46598764 | 0.56 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr13_+_30421472 | 0.56 |
ENSDART00000143569
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr16_+_46410520 | 0.56 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr24_-_5691956 | 0.54 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr19_-_31035325 | 0.53 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr15_+_24704798 | 0.52 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr24_-_21973163 | 0.52 |
ENSDART00000131406
|
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr8_-_38201415 | 0.51 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr9_-_12885201 | 0.51 |
ENSDART00000124957
|
ankzf1
|
ankyrin repeat and zinc finger domain containing 1 |
| chr18_-_15532016 | 0.51 |
ENSDART00000165279
|
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr22_+_4442473 | 0.50 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr17_+_21486047 | 0.50 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr2_+_25839650 | 0.50 |
ENSDART00000134077
ENSDART00000140804 |
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr18_+_44849809 | 0.50 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr2_+_25839940 | 0.49 |
ENSDART00000139927
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr9_+_28140089 | 0.49 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr11_+_19370447 | 0.48 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr17_+_23556764 | 0.47 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr14_+_20156477 | 0.47 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr5_+_65086856 | 0.46 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr22_+_17399124 | 0.45 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr1_+_46598502 | 0.45 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr5_+_65086668 | 0.45 |
ENSDART00000183746
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr22_+_23359369 | 0.45 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr14_+_3507326 | 0.45 |
ENSDART00000159326
|
gstp1
|
glutathione S-transferase pi 1 |
| chr5_-_20123002 | 0.45 |
ENSDART00000026516
|
pxmp2
|
peroxisomal membrane protein 2 |
| chr1_-_43905252 | 0.44 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr2_-_37277626 | 0.44 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr19_-_5805923 | 0.43 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr16_-_31475904 | 0.43 |
ENSDART00000145691
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr24_-_31090948 | 0.43 |
ENSDART00000176799
|
hccsb
|
holocytochrome c synthase b |
| chr12_-_17147473 | 0.43 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr20_-_35512932 | 0.42 |
ENSDART00000137690
|
adgrf3b
|
adhesion G protein-coupled receptor F3b |
| chr3_+_17616201 | 0.42 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr11_-_25461336 | 0.41 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr5_+_65087226 | 0.41 |
ENSDART00000183187
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr7_+_34549198 | 0.41 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr3_+_19687217 | 0.39 |
ENSDART00000141937
|
tlk2
|
tousled-like kinase 2 |
| chr7_+_7019911 | 0.39 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr18_-_41160579 | 0.38 |
ENSDART00000181480
|
CABZ01005876.1
|
|
| chr18_-_41160975 | 0.37 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr7_+_34549377 | 0.36 |
ENSDART00000191814
|
fhod1
|
formin homology 2 domain containing 1 |
| chr18_+_13077800 | 0.34 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr3_-_45250924 | 0.34 |
ENSDART00000109017
|
usp31
|
ubiquitin specific peptidase 31 |
| chr2_+_9822319 | 0.34 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr8_+_48965767 | 0.33 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr15_+_15771418 | 0.33 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr3_-_27646070 | 0.32 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr5_+_63767376 | 0.32 |
ENSDART00000138898
|
rgs3b
|
regulator of G protein signaling 3b |
| chr8_+_48966165 | 0.31 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr5_-_64355227 | 0.31 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr5_+_24089334 | 0.31 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr15_+_20352123 | 0.31 |
ENSDART00000011030
ENSDART00000163532 ENSDART00000169537 ENSDART00000161047 |
il15l
|
interleukin 15, like |
| chr13_-_33134611 | 0.30 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
| chr3_+_30921246 | 0.29 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr12_-_7639120 | 0.29 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr5_+_20035284 | 0.28 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr7_+_38770167 | 0.28 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr7_-_5396154 | 0.27 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr7_-_30926030 | 0.27 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr3_+_53352018 | 0.27 |
ENSDART00000082715
|
camsap3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr25_-_29072162 | 0.27 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr5_+_50953240 | 0.26 |
ENSDART00000148501
ENSDART00000149892 ENSDART00000190312 |
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr14_-_493029 | 0.23 |
ENSDART00000115093
|
zgc:172215
|
zgc:172215 |
| chr25_-_22187397 | 0.23 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr8_-_20838342 | 0.22 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr3_+_19216567 | 0.22 |
ENSDART00000134433
|
il12rb2l
|
interleukin 12 receptor, beta 2a, like |
| chr24_-_24797455 | 0.21 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr6_+_49901465 | 0.21 |
ENSDART00000023515
|
chmp4ba
|
charged multivesicular body protein 4Ba |
| chr7_-_19369002 | 0.21 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr5_-_54772982 | 0.21 |
ENSDART00000122829
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_-_24297510 | 0.20 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr14_+_30774032 | 0.20 |
ENSDART00000139552
|
atl3
|
atlastin 3 |
| chr5_+_52706316 | 0.20 |
ENSDART00000169488
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr20_+_32478151 | 0.18 |
ENSDART00000145269
|
ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr5_-_63644938 | 0.18 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr4_+_5317483 | 0.18 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chrM_+_8894 | 0.17 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr1_-_43933271 | 0.17 |
ENSDART00000121605
|
scpp6
|
secretory calcium-binding phosphoprotein 6 |
| chr1_+_52137528 | 0.16 |
ENSDART00000007079
ENSDART00000074265 |
asna1
|
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr4_-_77267787 | 0.16 |
ENSDART00000190346
|
zgc:174310
|
zgc:174310 |
| chr14_+_20900989 | 0.16 |
ENSDART00000106209
|
zgc:162941
|
zgc:162941 |
| chr25_+_16312258 | 0.15 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr4_+_7822773 | 0.15 |
ENSDART00000171391
|
si:ch1073-67j19.2
|
si:ch1073-67j19.2 |
| chr7_+_5530154 | 0.14 |
ENSDART00000181610
|
si:dkeyp-67a8.4
|
si:dkeyp-67a8.4 |
| chr21_-_11657043 | 0.13 |
ENSDART00000141297
|
cast
|
calpastatin |
| chr2_-_7605121 | 0.13 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr25_+_18583877 | 0.13 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr23_+_39462208 | 0.13 |
ENSDART00000136707
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr1_-_34335752 | 0.13 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr2_-_15362923 | 0.12 |
ENSDART00000166926
|
olfm3b
|
olfactomedin 3b |
| chr23_-_1017428 | 0.12 |
ENSDART00000110588
ENSDART00000183158 |
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr17_+_12942021 | 0.12 |
ENSDART00000192514
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr14_+_30774515 | 0.11 |
ENSDART00000191666
|
atl3
|
atlastin 3 |
| chr14_+_34486629 | 0.10 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr3_-_16493528 | 0.10 |
ENSDART00000142099
|
si:ch211-23l10.3
|
si:ch211-23l10.3 |
| chr22_-_11954861 | 0.10 |
ENSDART00000131611
|
si:dkeyp-4h4.1
|
si:dkeyp-4h4.1 |
| chr19_+_7001170 | 0.10 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr4_+_54899568 | 0.09 |
ENSDART00000162786
|
si:dkey-56m15.8
|
si:dkey-56m15.8 |
| chr12_+_23371542 | 0.09 |
ENSDART00000078896
|
waca
|
WW domain containing adaptor with coiled-coil a |
| chr18_+_23218980 | 0.09 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr17_+_33495194 | 0.09 |
ENSDART00000033691
|
pth2
|
parathyroid hormone 2 |
| chr3_+_33440615 | 0.09 |
ENSDART00000146005
|
gtpbp1
|
GTP binding protein 1 |
| chr3_+_16976095 | 0.09 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr23_+_39461989 | 0.09 |
ENSDART00000184761
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr20_-_38746889 | 0.09 |
ENSDART00000140275
|
trim54
|
tripartite motif containing 54 |
| chr19_+_4139065 | 0.09 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr25_+_32474031 | 0.08 |
ENSDART00000152124
|
sqor
|
sulfide quinone oxidoreductase |
| chr6_+_42475730 | 0.08 |
ENSDART00000150226
|
mst1ra
|
macrophage stimulating 1 receptor a |
| chr25_-_11378623 | 0.08 |
ENSDART00000166586
|
enc2
|
ectodermal-neural cortex 2 |
| chr21_+_40498628 | 0.08 |
ENSDART00000163454
|
coro6
|
coronin 6 |
| chr22_+_9862466 | 0.07 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr9_-_33062891 | 0.07 |
ENSDART00000161182
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr24_+_17140938 | 0.07 |
ENSDART00000149134
|
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr22_-_26595027 | 0.07 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr18_+_30421528 | 0.07 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| chrM_+_9052 | 0.07 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr6_+_55428924 | 0.07 |
ENSDART00000018270
|
ncoa5
|
nuclear receptor coactivator 5 |
| chr18_+_14342326 | 0.07 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr1_-_11689525 | 0.07 |
ENSDART00000090689
|
brat1
|
BRCA1-associated ATM activator 1 |
| chr13_-_40141630 | 0.07 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr25_+_8407892 | 0.06 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr8_-_14375890 | 0.06 |
ENSDART00000090306
|
xpr1a
|
xenotropic and polytropic retrovirus receptor 1a |
| chr4_+_43408004 | 0.06 |
ENSDART00000150476
|
si:dkeyp-53e4.2
|
si:dkeyp-53e4.2 |
| chr3_+_31177972 | 0.06 |
ENSDART00000185954
|
clec19a
|
C-type lectin domain containing 19A |
| chr21_+_3093419 | 0.06 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr8_-_11170114 | 0.06 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr8_+_44549290 | 0.06 |
ENSDART00000160801
|
si:dkey-220o5.5
|
si:dkey-220o5.5 |
| chr1_-_54765262 | 0.06 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr4_-_72476526 | 0.06 |
ENSDART00000174153
|
CR788316.1
|
|
| chr22_+_19218733 | 0.05 |
ENSDART00000183212
ENSDART00000133595 |
si:dkey-21e2.7
|
si:dkey-21e2.7 |
| chr2_-_42827336 | 0.05 |
ENSDART00000140913
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr4_-_69550757 | 0.05 |
ENSDART00000168344
|
znf1101
|
zinc finger protein 1101 |
| chr2_+_25278107 | 0.05 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr10_-_74408 | 0.05 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr5_-_14564878 | 0.05 |
ENSDART00000160511
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr20_+_23960525 | 0.05 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr2_-_6519017 | 0.05 |
ENSDART00000181716
|
rgs1
|
regulator of G protein signaling 1 |
| chr7_+_6317866 | 0.05 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr23_+_29570386 | 0.05 |
ENSDART00000193192
|
lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr20_-_35536668 | 0.05 |
ENSDART00000112866
|
adgrf11
|
adhesion G protein-coupled receptor F11 |
| chr14_-_31693148 | 0.05 |
ENSDART00000172958
|
map7d3
|
MAP7 domain containing 3 |
| chr18_-_19095141 | 0.05 |
ENSDART00000090962
ENSDART00000090922 |
dennd4a
|
DENN/MADD domain containing 4A |
| chr3_-_48259289 | 0.04 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr9_-_30346279 | 0.04 |
ENSDART00000089488
|
sytl5
|
synaptotagmin-like 5 |
| chr4_+_49196098 | 0.04 |
ENSDART00000150678
|
si:dkey-40n15.1
|
si:dkey-40n15.1 |
| chr4_+_55273988 | 0.04 |
ENSDART00000163388
|
si:dkey-157e10.8
|
si:dkey-157e10.8 |
| chr23_-_1017605 | 0.04 |
ENSDART00000138290
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr14_-_6936731 | 0.04 |
ENSDART00000162196
|
clk4a
|
CDC-like kinase 4a |
| chr4_-_72643171 | 0.04 |
ENSDART00000130126
|
CABZ01054394.1
|
|
| chr8_-_13177536 | 0.04 |
ENSDART00000100992
|
zgc:194990
|
zgc:194990 |
| chr25_-_13408760 | 0.03 |
ENSDART00000154445
|
gins3
|
GINS complex subunit 3 |
| chr23_+_25856541 | 0.03 |
ENSDART00000145426
ENSDART00000028236 |
hnf4a
|
hepatocyte nuclear factor 4, alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of jdp2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.3 | 0.9 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 0.8 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 0.5 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.2 | 0.5 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 1.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 1.0 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.3 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 1.7 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.4 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.7 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.6 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 1.3 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.3 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.5 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.1 | GO:0019418 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.3 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.3 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.8 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.0 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 1.1 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.5 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 1.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 0.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.1 | 0.5 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.6 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 1.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 1.0 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |