Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
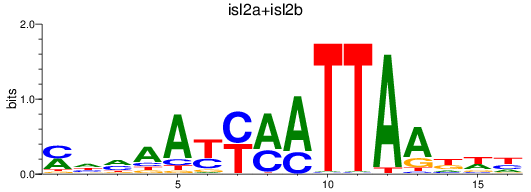
Results for isl2a+isl2b
Z-value: 0.54
Transcription factors associated with isl2a+isl2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl2a
|
ENSDARG00000003971 | ISL LIM homeobox 2a |
|
isl2b
|
ENSDARG00000053499 | ISL LIM homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl2a | dr11_v1_chr25_-_32751982_32751982 | 0.92 | 8.9e-08 | Click! |
| isl2b | dr11_v1_chr7_+_29992889_29992889 | 0.84 | 1.5e-05 | Click! |
Activity profile of isl2a+isl2b motif
Sorted Z-values of isl2a+isl2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_15433518 | 2.60 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr17_+_15433671 | 2.33 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr15_+_34988148 | 2.07 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr10_-_17083180 | 1.63 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr24_+_33622769 | 1.61 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr5_-_41494831 | 1.51 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr5_+_69747417 | 1.37 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr3_-_19200571 | 1.36 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr8_+_24747865 | 1.33 |
ENSDART00000078656
|
slc16a4
|
solute carrier family 16, member 4 |
| chr3_-_39180048 | 1.25 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr20_+_25586099 | 1.00 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr17_-_12407106 | 1.00 |
ENSDART00000183540
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr23_-_36724575 | 0.97 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr8_-_13046089 | 0.96 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr16_+_10972733 | 0.95 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr3_+_14388010 | 0.86 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr7_+_39166460 | 0.85 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr21_+_11385031 | 0.84 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr24_-_26310854 | 0.80 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr18_-_48547564 | 0.77 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr24_+_12989727 | 0.74 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr2_-_293793 | 0.73 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr25_+_21833287 | 0.73 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr23_-_16485190 | 0.71 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr2_+_19522082 | 0.69 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr19_+_31904836 | 0.69 |
ENSDART00000162297
ENSDART00000088340 ENSDART00000151280 ENSDART00000151218 |
tpd52
|
tumor protein D52 |
| chr6_-_8311044 | 0.68 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr6_-_49510553 | 0.67 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr8_-_50147948 | 0.65 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr16_+_28728347 | 0.65 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr13_-_9450210 | 0.63 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr19_+_1688727 | 0.63 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr4_-_9909371 | 0.61 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr14_-_24391424 | 0.60 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr2_+_19633493 | 0.60 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr7_+_22657566 | 0.59 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr20_-_23171430 | 0.58 |
ENSDART00000109234
|
spata18
|
spermatogenesis associated 18 |
| chr13_+_9046473 | 0.58 |
ENSDART00000136557
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr20_+_11731039 | 0.58 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr6_+_9204099 | 0.57 |
ENSDART00000150167
ENSDART00000115394 |
tbx19
|
T-box 19 |
| chr6_+_52350443 | 0.57 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr3_-_30061985 | 0.55 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr2_+_19578446 | 0.55 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr7_+_26534131 | 0.54 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr2_-_293036 | 0.54 |
ENSDART00000171629
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr17_+_12285285 | 0.52 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr21_-_19314618 | 0.51 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr13_-_9467944 | 0.50 |
ENSDART00000136582
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr1_-_669717 | 0.50 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr13_-_36391496 | 0.50 |
ENSDART00000100217
ENSDART00000140243 |
actn1
|
actinin, alpha 1 |
| chr23_+_39695827 | 0.47 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr15_-_14884332 | 0.47 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr15_-_46779934 | 0.47 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr3_-_13068189 | 0.46 |
ENSDART00000167180
|
prkar1b
|
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_-_52210950 | 0.46 |
ENSDART00000083946
|
pld6
|
phospholipase D family, member 6 |
| chr20_+_40457599 | 0.46 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr7_-_48665305 | 0.46 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr8_-_52229462 | 0.46 |
ENSDART00000185949
ENSDART00000051825 |
tcf7l1b
|
transcription factor 7 like 1b |
| chr20_-_47188966 | 0.45 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr18_-_15932704 | 0.44 |
ENSDART00000127769
|
plekhg7
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
| chr1_-_42289704 | 0.43 |
ENSDART00000150124
|
si:ch211-71k14.1
|
si:ch211-71k14.1 |
| chr16_-_21140097 | 0.43 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr22_-_5252005 | 0.41 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr2_-_19520690 | 0.41 |
ENSDART00000133559
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr21_+_45841731 | 0.40 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr11_+_30057762 | 0.39 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr8_-_7475917 | 0.39 |
ENSDART00000082157
|
gata1b
|
GATA binding protein 1b |
| chr2_-_19520324 | 0.39 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr10_-_17222083 | 0.39 |
ENSDART00000134059
|
depdc5
|
DEP domain containing 5 |
| chr21_-_20939488 | 0.39 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr13_+_646700 | 0.38 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr8_+_52637507 | 0.38 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr19_+_9186175 | 0.38 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr4_-_17669881 | 0.37 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr24_-_12770357 | 0.37 |
ENSDART00000060826
|
ipo4
|
importin 4 |
| chr17_-_14815557 | 0.36 |
ENSDART00000154473
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr11_-_45138857 | 0.35 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr14_-_7137808 | 0.35 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr11_-_1509773 | 0.35 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr23_+_4253957 | 0.34 |
ENSDART00000008761
|
arl6ip5b
|
ADP-ribosylation factor-like 6 interacting protein 5b |
| chr1_-_50859053 | 0.34 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr24_+_13869092 | 0.32 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr11_+_45436703 | 0.32 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr15_+_9327252 | 0.32 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr23_-_18024543 | 0.29 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr25_+_7671640 | 0.29 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr22_-_36530902 | 0.28 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr25_+_13406069 | 0.28 |
ENSDART00000010495
|
znrf1
|
zinc and ring finger 1 |
| chr7_+_59020972 | 0.28 |
ENSDART00000157873
|
hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr24_-_6078222 | 0.28 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr12_+_24342303 | 0.28 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr18_-_8398972 | 0.28 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr6_+_54538948 | 0.28 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr22_+_18816662 | 0.26 |
ENSDART00000132476
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr17_-_29224908 | 0.26 |
ENSDART00000156288
|
si:dkey-28g23.6
|
si:dkey-28g23.6 |
| chr16_-_17072440 | 0.26 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr12_-_4781801 | 0.25 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr11_-_1400507 | 0.25 |
ENSDART00000173029
ENSDART00000172953 ENSDART00000111140 |
rpl29
|
ribosomal protein L29 |
| chr14_-_413273 | 0.25 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr12_-_6880694 | 0.25 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr23_-_18913032 | 0.24 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr12_-_28363111 | 0.24 |
ENSDART00000016283
ENSDART00000164156 |
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr3_+_38540411 | 0.24 |
ENSDART00000154943
|
si:dkey-7f16.3
|
si:dkey-7f16.3 |
| chr22_+_19552987 | 0.24 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr16_-_48400639 | 0.22 |
ENSDART00000159372
|
eif3ha
|
eukaryotic translation initiation factor 3, subunit H, a |
| chr1_+_55608520 | 0.22 |
ENSDART00000152307
|
adgre18
|
adhesion G protein-coupled receptor E18 |
| chr2_-_22927958 | 0.21 |
ENSDART00000141621
|
myo7bb
|
myosin VIIBb |
| chr11_-_29737088 | 0.21 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr12_+_18899396 | 0.21 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr15_-_21702317 | 0.19 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr16_-_45094599 | 0.19 |
ENSDART00000155479
|
si:rp71-77l1.1
|
si:rp71-77l1.1 |
| chr4_-_75158035 | 0.18 |
ENSDART00000174353
|
CABZ01066312.1
|
|
| chr9_+_51147380 | 0.18 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr22_-_19552796 | 0.17 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr8_+_1651821 | 0.16 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr23_+_32406461 | 0.16 |
ENSDART00000179878
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr17_+_43595692 | 0.16 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr16_+_23303859 | 0.15 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
| chr12_+_18681477 | 0.15 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr3_+_32553714 | 0.15 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr25_+_35212919 | 0.15 |
ENSDART00000180127
|
ano5a
|
anoctamin 5a |
| chr2_+_10771787 | 0.14 |
ENSDART00000187782
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr10_-_43771447 | 0.14 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr3_-_22216771 | 0.14 |
ENSDART00000130546
|
maptb
|
microtubule-associated protein tau b |
| chr1_-_10048514 | 0.14 |
ENSDART00000125358
ENSDART00000054835 |
rnf175
|
ring finger protein 175 |
| chr14_-_4145594 | 0.14 |
ENSDART00000077348
|
casp3b
|
caspase 3, apoptosis-related cysteine peptidase b |
| chr13_-_12602920 | 0.14 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr22_-_17482282 | 0.13 |
ENSDART00000105453
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr23_+_9353552 | 0.13 |
ENSDART00000163298
|
BX511246.1
|
|
| chr1_-_55873178 | 0.12 |
ENSDART00000019936
|
prkacab
|
protein kinase, cAMP-dependent, catalytic, alpha, genome duplicate b |
| chr8_-_53926228 | 0.12 |
ENSDART00000015554
|
ubiad1
|
UbiA prenyltransferase domain containing 1 |
| chr6_+_39905021 | 0.12 |
ENSDART00000064904
|
endou
|
endonuclease, polyU-specific |
| chr5_-_19861766 | 0.12 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr21_-_35419486 | 0.12 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr23_+_32426954 | 0.12 |
ENSDART00000192529
ENSDART00000172313 |
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr10_-_7756865 | 0.11 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr21_-_37194839 | 0.11 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr3_-_36839115 | 0.10 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr7_+_66822229 | 0.10 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr1_+_51721851 | 0.10 |
ENSDART00000040397
|
prdx2
|
peroxiredoxin 2 |
| chr19_+_24872159 | 0.10 |
ENSDART00000158490
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr15_-_14552101 | 0.10 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr15_+_36115955 | 0.09 |
ENSDART00000032702
|
sst1.2
|
somatostatin 1, tandem duplicate 2 |
| chr22_-_17844117 | 0.09 |
ENSDART00000159363
|
BX908731.1
|
|
| chr20_-_5291012 | 0.09 |
ENSDART00000122892
|
cyp46a1.3
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 3 |
| chr6_+_9175886 | 0.08 |
ENSDART00000165333
|
si:ch211-207l14.1
|
si:ch211-207l14.1 |
| chr11_-_20956309 | 0.08 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr3_-_19368435 | 0.08 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr4_-_77218637 | 0.08 |
ENSDART00000174325
|
psmb10
|
proteasome subunit beta 10 |
| chr13_-_42560662 | 0.08 |
ENSDART00000124898
|
CR792417.1
|
|
| chr19_+_16015881 | 0.07 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr13_+_7049823 | 0.07 |
ENSDART00000178997
ENSDART00000161443 |
rnaset2
|
ribonuclease T2 |
| chr3_-_25119839 | 0.07 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr8_-_22273819 | 0.07 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr11_+_29856032 | 0.07 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr5_+_58550291 | 0.07 |
ENSDART00000184983
ENSDART00000044803 |
pou2f3
|
POU class 2 homeobox 3 |
| chr18_-_3473081 | 0.07 |
ENSDART00000174975
|
myo7aa
|
myosin VIIAa |
| chr14_-_4682114 | 0.06 |
ENSDART00000014454
|
gabra4
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 4 |
| chr17_-_24353462 | 0.06 |
ENSDART00000191021
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr21_-_35534401 | 0.06 |
ENSDART00000112308
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr16_+_28932038 | 0.06 |
ENSDART00000149480
|
npr1b
|
natriuretic peptide receptor 1b |
| chr7_+_20512419 | 0.06 |
ENSDART00000173907
|
si:dkey-19b23.14
|
si:dkey-19b23.14 |
| chr2_-_59205393 | 0.05 |
ENSDART00000056417
ENSDART00000182452 ENSDART00000141876 |
ftr30
|
finTRIM family, member 30 |
| chr5_+_56023186 | 0.05 |
ENSDART00000156230
|
fzd9a
|
frizzled class receptor 9a |
| chr19_-_5125663 | 0.04 |
ENSDART00000005547
ENSDART00000132642 |
gnb3b
|
guanine nucleotide binding protein (G protein), beta polypeptide 3b |
| chr12_+_16168342 | 0.04 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr8_+_25913787 | 0.04 |
ENSDART00000190257
ENSDART00000062515 |
sema3h
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3H |
| chr19_+_45962016 | 0.04 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr4_-_49582758 | 0.04 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr2_-_45027754 | 0.04 |
ENSDART00000168469
|
vwa5b2
|
von Willebrand factor A domain containing 5B2 |
| chr22_-_17482810 | 0.03 |
ENSDART00000171293
|
si:ch211-197g15.7
|
si:ch211-197g15.7 |
| chr18_+_30847237 | 0.03 |
ENSDART00000012374
|
foxf1
|
forkhead box F1 |
| chr6_-_16717878 | 0.03 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr6_-_54486410 | 0.03 |
ENSDART00000064679
|
CR847944.1
|
|
| chr7_-_12821277 | 0.03 |
ENSDART00000091584
|
zgc:158785
|
zgc:158785 |
| chr23_-_33350990 | 0.03 |
ENSDART00000144831
|
si:ch211-226m16.2
|
si:ch211-226m16.2 |
| chr5_-_42083363 | 0.03 |
ENSDART00000162596
|
cxcl11.5
|
chemokine (C-X-C motif) ligand 11, duplicate 5 |
| chr5_+_22791686 | 0.02 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr10_+_40568735 | 0.02 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr6_-_35046735 | 0.02 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr6_-_12588044 | 0.02 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr9_+_24008879 | 0.02 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr19_-_20403845 | 0.02 |
ENSDART00000151265
ENSDART00000147911 ENSDART00000151356 |
dazl
|
deleted in azoospermia-like |
| chr3_+_3681116 | 0.02 |
ENSDART00000109618
|
art4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr20_-_46128590 | 0.02 |
ENSDART00000123744
|
taar1b
|
trace amine associated receptor 1b |
| chr1_+_55279124 | 0.02 |
ENSDART00000152660
|
si:ch211-286b5.6
|
si:ch211-286b5.6 |
| chr22_-_23545307 | 0.02 |
ENSDART00000166915
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr12_-_34713690 | 0.01 |
ENSDART00000180807
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr16_-_16761164 | 0.01 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr13_+_38430466 | 0.01 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr20_+_66857 | 0.01 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr10_-_40964739 | 0.01 |
ENSDART00000076359
|
npffr1l1
|
neuropeptide FF receptor 1 like 1 |
| chr4_-_2219705 | 0.01 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr5_-_31901468 | 0.01 |
ENSDART00000147814
ENSDART00000141446 |
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr22_-_23545828 | 0.00 |
ENSDART00000161253
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr14_-_30932373 | 0.00 |
ENSDART00000172988
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr22_-_12745589 | 0.00 |
ENSDART00000136841
|
plcd4a
|
phospholipase C, delta 4a |
| chr11_+_30663300 | 0.00 |
ENSDART00000161662
|
ttbk1a
|
tau tubulin kinase 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl2a+isl2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.5 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.1 | 0.3 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.8 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.4 | GO:2000381 | negative regulation of nodal signaling pathway(GO:1900108) negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.4 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.7 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.5 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.6 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.8 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.3 | GO:1903306 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.3 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 2.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.7 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.5 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.8 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 4.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.3 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.0 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 1.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0043202 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.5 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.0 | GO:0097268 | cytoophidium(GO:0097268) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 0.8 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.2 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.5 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.7 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 1.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.4 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.6 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.5 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |