Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
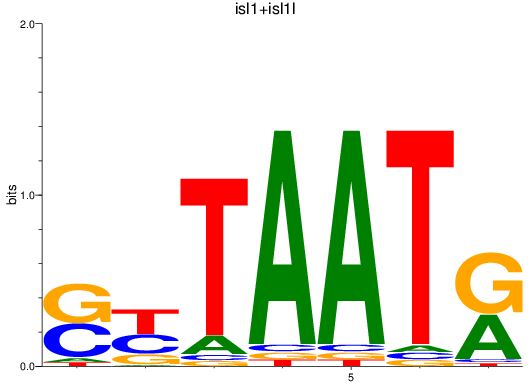
Results for isl1+isl1l
Z-value: 0.36
Transcription factors associated with isl1+isl1l
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
isl1
|
ENSDARG00000004023 | ISL LIM homeobox 1 |
|
isl1l
|
ENSDARG00000021055 | islet1, like |
|
isl1l
|
ENSDARG00000110204 | islet1, like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| isl1 | dr11_v1_chr5_-_40734045_40734045 | -0.93 | 3.0e-08 | Click! |
| isl1l | dr11_v1_chr10_+_8680730_8680730 | -0.83 | 2.3e-05 | Click! |
Activity profile of isl1+isl1l motif
Sorted Z-values of isl1+isl1l motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_29387215 | 1.45 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr9_-_35633827 | 1.04 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr12_+_22672323 | 0.97 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr2_-_17114852 | 0.91 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr20_-_38787341 | 0.87 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr16_+_30117798 | 0.83 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr20_-_45772306 | 0.81 |
ENSDART00000062092
|
trmt6
|
tRNA methyltransferase 6 homolog (S. cerevisiae) |
| chr13_+_46941930 | 0.80 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr25_+_36292057 | 0.75 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr20_-_38787047 | 0.74 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr17_-_4252221 | 0.73 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr22_-_17653143 | 0.66 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr1_-_9486214 | 0.66 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr13_+_22712406 | 0.65 |
ENSDART00000132847
|
si:ch211-134m17.9
|
si:ch211-134m17.9 |
| chr10_-_2943474 | 0.60 |
ENSDART00000188698
|
oclna
|
occludin a |
| chr8_-_51367298 | 0.59 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr4_+_26056548 | 0.59 |
ENSDART00000171204
|
SCYL2
|
si:ch211-244b2.1 |
| chr23_+_19590598 | 0.59 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr12_-_10505986 | 0.58 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr20_-_48407944 | 0.57 |
ENSDART00000085624
|
ipo13
|
importin 13 |
| chr6_+_10338554 | 0.55 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr24_-_33308045 | 0.51 |
ENSDART00000149711
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr8_-_25033681 | 0.51 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr11_-_279328 | 0.50 |
ENSDART00000066179
|
npffl
|
neuropeptide FF-amide peptide precursor like |
| chr11_+_24316110 | 0.50 |
ENSDART00000186028
|
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr21_-_35325466 | 0.50 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr19_-_47570672 | 0.49 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_-_36597612 | 0.49 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr3_+_24062748 | 0.49 |
ENSDART00000188574
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr25_+_7492663 | 0.48 |
ENSDART00000166496
|
cat
|
catalase |
| chr22_-_3299100 | 0.47 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr21_-_21178410 | 0.46 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr14_-_41478265 | 0.45 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr25_+_7532627 | 0.45 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr10_+_26990095 | 0.43 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr23_-_45398622 | 0.43 |
ENSDART00000053571
ENSDART00000149464 |
zgc:100911
|
zgc:100911 |
| chr25_+_35889102 | 0.42 |
ENSDART00000023453
ENSDART00000125821 ENSDART00000135441 |
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr22_-_28644517 | 0.42 |
ENSDART00000189538
|
jagn1b
|
jagunal homolog 1b |
| chr23_+_20431388 | 0.42 |
ENSDART00000132920
ENSDART00000102963 ENSDART00000109899 ENSDART00000140219 |
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr14_+_16287968 | 0.41 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr23_+_20431140 | 0.40 |
ENSDART00000193950
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr17_-_29271359 | 0.39 |
ENSDART00000104219
|
rcor1
|
REST corepressor 1 |
| chr4_-_837768 | 0.39 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr18_+_3243292 | 0.39 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr8_+_21254192 | 0.39 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr14_-_30876299 | 0.39 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr4_-_12930086 | 0.39 |
ENSDART00000013604
|
lemd3
|
LEM domain containing 3 |
| chr21_+_31253048 | 0.38 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr6_-_53326421 | 0.38 |
ENSDART00000191740
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr12_+_13205955 | 0.38 |
ENSDART00000092906
|
ppp1cab
|
protein phosphatase 1, catalytic subunit, alpha isozyme b |
| chr2_-_1622641 | 0.37 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr11_+_45110865 | 0.36 |
ENSDART00000158188
|
mgea5l
|
meningioma expressed antigen 5 (hyaluronidase) like |
| chr25_+_6122823 | 0.36 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr22_+_438714 | 0.35 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr7_-_71585065 | 0.35 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr18_-_12295092 | 0.35 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr15_-_43625549 | 0.34 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr15_-_23529945 | 0.34 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr19_+_42432625 | 0.34 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr10_-_1961576 | 0.33 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr19_-_6193448 | 0.33 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr20_+_29209926 | 0.33 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr12_+_14149686 | 0.33 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr3_-_1283247 | 0.32 |
ENSDART00000149814
|
tcf20
|
transcription factor 20 |
| chr6_-_39005133 | 0.32 |
ENSDART00000104116
ENSDART00000151750 |
vdrb
|
vitamin D receptor b |
| chr15_-_44052927 | 0.32 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr8_-_30338872 | 0.31 |
ENSDART00000137583
|
dock8
|
dedicator of cytokinesis 8 |
| chr5_+_31168096 | 0.31 |
ENSDART00000086443
ENSDART00000192271 |
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr6_-_31682135 | 0.31 |
ENSDART00000153988
|
cachd1
|
cache domain containing 1 |
| chr9_-_41507712 | 0.30 |
ENSDART00000135821
|
mfsd6b
|
major facilitator superfamily domain containing 6b |
| chr8_+_48942470 | 0.30 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr25_-_10769039 | 0.30 |
ENSDART00000186758
ENSDART00000121724 |
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr4_-_13156971 | 0.29 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr14_-_36437249 | 0.29 |
ENSDART00000016728
|
aga
|
aspartylglucosaminidase |
| chr23_-_32156278 | 0.28 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr23_+_31596441 | 0.28 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr23_+_2703044 | 0.28 |
ENSDART00000182512
ENSDART00000105286 |
ncoa6
|
nuclear receptor coactivator 6 |
| chr5_-_13076779 | 0.28 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
| chr20_+_18703951 | 0.28 |
ENSDART00000191070
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr4_+_9478500 | 0.27 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr16_-_55259199 | 0.27 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr11_+_30647545 | 0.27 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr21_+_26522571 | 0.27 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr6_-_33707278 | 0.27 |
ENSDART00000188103
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr9_+_41459759 | 0.26 |
ENSDART00000132501
ENSDART00000100265 |
nemp2
|
nuclear envelope integral membrane protein 2 |
| chr25_+_32390794 | 0.26 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr17_+_47090497 | 0.26 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr19_+_8612839 | 0.26 |
ENSDART00000144925
|
snx27a
|
sorting nexin family member 27a |
| chr17_-_13070602 | 0.26 |
ENSDART00000188311
ENSDART00000193428 |
CU469462.1
|
|
| chr12_-_13205854 | 0.26 |
ENSDART00000077829
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr3_-_23513155 | 0.25 |
ENSDART00000170200
|
BX682558.1
|
|
| chr5_+_66250856 | 0.25 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr9_-_14137295 | 0.25 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr17_+_33375469 | 0.24 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr7_-_30280934 | 0.24 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr16_+_52999778 | 0.24 |
ENSDART00000011506
|
nkd2a
|
naked cuticle homolog 2a |
| chr3_-_18737126 | 0.24 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr5_-_19006290 | 0.24 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr11_-_31226578 | 0.23 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr3_+_13860815 | 0.23 |
ENSDART00000162451
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr8_+_52530889 | 0.23 |
ENSDART00000127729
ENSDART00000170360 ENSDART00000162687 |
stambpb
|
STAM binding protein b |
| chr10_-_17587832 | 0.23 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr20_-_40360571 | 0.23 |
ENSDART00000144768
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr23_-_36934944 | 0.23 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr5_+_25762271 | 0.23 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr8_-_53896982 | 0.22 |
ENSDART00000169206
|
MIB2
|
mindbomb E3 ubiquitin protein ligase 2 |
| chr25_+_30131055 | 0.22 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr11_-_14813029 | 0.22 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr20_+_25712276 | 0.22 |
ENSDART00000121585
ENSDART00000185772 |
cep135
|
centrosomal protein 135 |
| chr5_+_31791001 | 0.22 |
ENSDART00000043010
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
| chr5_-_33281046 | 0.22 |
ENSDART00000051344
ENSDART00000138116 |
surf6
|
surfeit 6 |
| chr2_+_1881334 | 0.22 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr16_+_9400661 | 0.22 |
ENSDART00000146174
|
ice1
|
KIAA0947-like (H. sapiens) |
| chr10_-_13343831 | 0.21 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr16_+_45337663 | 0.21 |
ENSDART00000147343
ENSDART00000185731 |
dnase2
|
deoxyribonuclease II, lysosomal |
| chr11_+_16216909 | 0.21 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr11_-_18017287 | 0.20 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr1_+_30946231 | 0.20 |
ENSDART00000022841
|
metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr20_+_25711718 | 0.20 |
ENSDART00000033436
|
cep135
|
centrosomal protein 135 |
| chr12_-_13205572 | 0.20 |
ENSDART00000152670
|
pelo
|
pelota mRNA surveillance and ribosome rescue factor |
| chr6_-_49898881 | 0.20 |
ENSDART00000150204
|
atp5f1e
|
ATP synthase F1 subunit epsilon |
| chr4_-_27897160 | 0.20 |
ENSDART00000066924
ENSDART00000066925 ENSDART00000193020 |
tbc1d22a
|
TBC1 domain family, member 22a |
| chr25_+_16098620 | 0.20 |
ENSDART00000142564
ENSDART00000165598 |
far1
|
fatty acyl CoA reductase 1 |
| chr1_-_25144439 | 0.20 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr10_-_10018120 | 0.20 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr8_-_22838364 | 0.20 |
ENSDART00000187954
ENSDART00000185368 ENSDART00000188210 |
magixa
|
MAGI family member, X-linked a |
| chr11_+_41459408 | 0.19 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr15_+_23657051 | 0.19 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr16_+_13883872 | 0.19 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr10_+_7593185 | 0.19 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr15_+_11814969 | 0.19 |
ENSDART00000127248
|
FO704748.1
|
|
| chr4_-_15103646 | 0.19 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr1_-_23596391 | 0.18 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr7_+_36898850 | 0.18 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr6_-_19333947 | 0.17 |
ENSDART00000160887
|
gga3
|
ggolgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr16_-_36979592 | 0.17 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr19_-_7272921 | 0.17 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr14_+_23709134 | 0.17 |
ENSDART00000191162
ENSDART00000179754 ENSDART00000054266 |
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr13_-_48388726 | 0.17 |
ENSDART00000169473
|
fbxo11a
|
F-box protein 11a |
| chr11_-_18107447 | 0.17 |
ENSDART00000187376
|
qrich1
|
glutamine-rich 1 |
| chr10_+_35352435 | 0.17 |
ENSDART00000123936
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr22_-_16755885 | 0.17 |
ENSDART00000036467
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr11_-_10850936 | 0.16 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr6_+_36844127 | 0.16 |
ENSDART00000020505
|
chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr7_-_34192834 | 0.16 |
ENSDART00000125131
|
smad6a
|
SMAD family member 6a |
| chr2_+_45100963 | 0.15 |
ENSDART00000160122
|
CU929150.2
|
|
| chr21_+_2506013 | 0.15 |
ENSDART00000162351
|
hmgcrb
|
3-hydroxy-3-methylglutaryl-CoA reductase b |
| chr25_+_459901 | 0.15 |
ENSDART00000154895
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr22_+_26853254 | 0.15 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr3_-_11008532 | 0.15 |
ENSDART00000165086
|
CR382337.3
|
|
| chr18_+_35861930 | 0.15 |
ENSDART00000185223
|
ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr7_-_54679595 | 0.15 |
ENSDART00000165320
|
ccnd1
|
cyclin D1 |
| chr25_-_32381642 | 0.15 |
ENSDART00000133872
ENSDART00000006124 |
cops2
|
COP9 signalosome subunit 2 |
| chr21_+_34981263 | 0.15 |
ENSDART00000132711
|
rbm11
|
RNA binding motif protein 11 |
| chr2_-_31711547 | 0.14 |
ENSDART00000175525
ENSDART00000189763 ENSDART00000113498 |
lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr9_+_37320222 | 0.14 |
ENSDART00000100622
|
slc15a2
|
solute carrier family 15 (oligopeptide transporter), member 2 |
| chr8_+_20140321 | 0.14 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr3_-_58650057 | 0.14 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr9_-_41025062 | 0.14 |
ENSDART00000002053
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr12_+_27034760 | 0.14 |
ENSDART00000181170
ENSDART00000153054 |
fbrs
|
fibrosin |
| chr15_+_1199407 | 0.13 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr7_+_67748939 | 0.13 |
ENSDART00000162978
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr20_-_5400395 | 0.13 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr17_-_36529016 | 0.13 |
ENSDART00000025019
|
colec11
|
collectin sub-family member 11 |
| chr13_-_15082024 | 0.13 |
ENSDART00000157482
|
sfxn5a
|
sideroflexin 5a |
| chr4_+_2230701 | 0.13 |
ENSDART00000080439
|
cmah
|
cytidine monophospho-N-acetylneuraminic acid hydroxylase |
| chr18_-_14743659 | 0.13 |
ENSDART00000125979
|
tshz3a
|
teashirt zinc finger homeobox 3a |
| chr13_+_31184796 | 0.12 |
ENSDART00000147085
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr7_+_67429185 | 0.12 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr14_-_32631013 | 0.11 |
ENSDART00000176815
|
atp11c
|
ATPase phospholipid transporting 11C |
| chr23_-_28423048 | 0.11 |
ENSDART00000140448
|
c1ql4a
|
complement component 1, q subcomponent-like 4 |
| chr23_-_351935 | 0.11 |
ENSDART00000140986
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| chr5_-_56948058 | 0.11 |
ENSDART00000083074
ENSDART00000191028 |
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr12_+_27034594 | 0.11 |
ENSDART00000111679
|
fbrs
|
fibrosin |
| chr13_-_9159852 | 0.11 |
ENSDART00000170185
ENSDART00000158697 ENSDART00000143393 ENSDART00000164973 ENSDART00000159910 |
HTRA2 (1 of many)
|
si:dkey-112g5.16 |
| chr15_+_888704 | 0.11 |
ENSDART00000182796
|
si:dkey-7i4.9
|
si:dkey-7i4.9 |
| chr3_-_25420931 | 0.11 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr16_-_35415004 | 0.11 |
ENSDART00000170401
ENSDART00000157832 ENSDART00000170048 |
taf12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr13_+_16522608 | 0.11 |
ENSDART00000182838
ENSDART00000143200 |
kcnma1a
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1a |
| chr17_-_2036850 | 0.10 |
ENSDART00000186048
ENSDART00000188838 ENSDART00000186423 |
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr3_-_25148047 | 0.10 |
ENSDART00000089325
|
mief1
|
mitochondrial elongation factor 1 |
| chr15_-_24588649 | 0.10 |
ENSDART00000060469
|
gemin4
|
gem (nuclear organelle) associated protein 4 |
| chr7_+_15677196 | 0.10 |
ENSDART00000192676
|
mef2ab
|
myocyte enhancer factor 2ab |
| chr9_+_8942258 | 0.10 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr13_+_31177934 | 0.10 |
ENSDART00000144568
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr9_-_27720612 | 0.10 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr8_+_23658626 | 0.10 |
ENSDART00000083605
|
tbc1d25
|
TBC1 domain family, member 25 |
| chr10_+_35358675 | 0.10 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr11_-_35975026 | 0.10 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr22_-_7563593 | 0.10 |
ENSDART00000106062
|
BX511034.1
|
|
| chr21_-_30408775 | 0.09 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr22_-_31020690 | 0.09 |
ENSDART00000130604
|
ssuh2.4
|
ssu-2 homolog, tandem duplicate 4 |
| chr15_-_20731297 | 0.09 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr3_-_21062706 | 0.09 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr19_+_9786722 | 0.09 |
ENSDART00000138310
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr8_+_54055390 | 0.09 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr22_+_31023205 | 0.08 |
ENSDART00000111561
|
zmp:0000000735
|
zmp:0000000735 |
| chr12_+_10062953 | 0.08 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr4_+_17539134 | 0.08 |
ENSDART00000013313
|
itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr17_+_44756247 | 0.08 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
Network of associatons between targets according to the STRING database.
First level regulatory network of isl1+isl1l
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 0.8 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.4 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.3 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.1 | 0.4 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.3 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 0.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.9 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 0.3 | GO:0019884 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.2 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.4 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0031645 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.4 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.2 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 0.3 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.3 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 1.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.3 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.1 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.3 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.2 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.3 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.1 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.0 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.6 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.1 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) |
| 0.0 | 0.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.5 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.5 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.6 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.0 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.0 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.1 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0090104 | pancreatic D cell differentiation(GO:0003311) pancreatic epsilon cell differentiation(GO:0090104) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 0.4 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.0 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.2 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 0.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.5 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.3 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 1.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.0 | 0.4 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.4 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.4 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |