Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
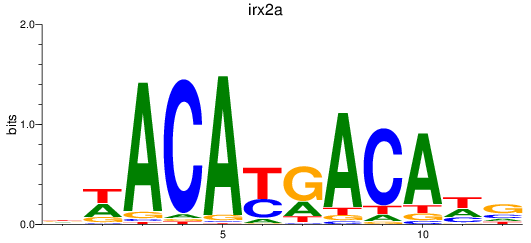
Results for irx2a
Z-value: 0.38
Transcription factors associated with irx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx2a
|
ENSDARG00000001785 | iroquois homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx2a | dr11_v1_chr16_-_563732_563732 | 0.26 | 3.0e-01 | Click! |
Activity profile of irx2a motif
Sorted Z-values of irx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_17083180 | 1.35 |
ENSDART00000170083
|
fam166b
|
family with sequence similarity 166, member B |
| chr12_-_11649690 | 1.15 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr5_-_22621417 | 1.00 |
ENSDART00000138716
|
zgc:113208
|
zgc:113208 |
| chr12_+_2870671 | 0.99 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr18_-_29961784 | 0.88 |
ENSDART00000135601
ENSDART00000140661 |
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr12_+_36891483 | 0.86 |
ENSDART00000048927
|
cox10
|
COX10 heme A:farnesyltransferase cytochrome c oxidase assembly factor |
| chr1_+_55239710 | 0.82 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr13_-_11971148 | 0.80 |
ENSDART00000066230
ENSDART00000185614 |
ARL3 (1 of many)
|
zgc:110197 |
| chr23_-_28141419 | 0.78 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr23_+_42819221 | 0.72 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr3_+_55105066 | 0.72 |
ENSDART00000110848
|
si:ch211-5k11.8
|
si:ch211-5k11.8 |
| chr19_-_41069573 | 0.70 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr14_-_33360344 | 0.67 |
ENSDART00000181291
|
pimr117
|
Pim proto-oncogene, serine/threonine kinase, related 117 |
| chr22_-_21314821 | 0.66 |
ENSDART00000105546
ENSDART00000135388 |
cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr12_+_10163585 | 0.62 |
ENSDART00000106191
|
psmc5
|
proteasome 26S subunit, ATPase 5 |
| chr6_-_24320268 | 0.54 |
ENSDART00000171733
|
si:dkeyp-85h7.1
|
si:dkeyp-85h7.1 |
| chr6_-_54845622 | 0.52 |
ENSDART00000048887
|
csrp1b
|
cysteine and glycine-rich protein 1b |
| chr19_+_9004256 | 0.51 |
ENSDART00000186401
ENSDART00000091443 |
si:ch211-81a5.1
|
si:ch211-81a5.1 |
| chr21_+_22423286 | 0.50 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr5_+_338154 | 0.49 |
ENSDART00000191743
|
rnf170
|
ring finger protein 170 |
| chr14_-_48103207 | 0.48 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr6_-_7720332 | 0.47 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr24_-_37955993 | 0.46 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr8_+_26565512 | 0.45 |
ENSDART00000140980
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr14_-_26400501 | 0.45 |
ENSDART00000054189
|
pimr212
|
Pim proto-oncogene, serine/threonine kinase, related 212 |
| chr24_-_7632187 | 0.44 |
ENSDART00000041714
|
atp6v0a1b
|
ATPase H+ transporting V0 subunit a1b |
| chr23_+_4299887 | 0.44 |
ENSDART00000132604
|
l3mbtl1a
|
l(3)mbt-like 1a (Drosophila) |
| chr21_+_723998 | 0.42 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr13_+_32636197 | 0.42 |
ENSDART00000141057
|
si:ch211-206k20.5
|
si:ch211-206k20.5 |
| chr22_+_7480465 | 0.41 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr12_+_27024676 | 0.41 |
ENSDART00000153104
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr7_+_17255705 | 0.40 |
ENSDART00000053357
ENSDART00000167849 ENSDART00000165833 |
nitr9
si:ch73-46n24.1
|
novel immune-type receptor 9 si:ch73-46n24.1 |
| chr23_+_23658474 | 0.39 |
ENSDART00000162838
|
agrn
|
agrin |
| chr21_-_37733287 | 0.39 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr1_-_49544601 | 0.39 |
ENSDART00000145960
|
slc47a2.2
|
solute carrier family 47 (multidrug and toxin extrusion), member 2.2 |
| chr2_+_44545912 | 0.38 |
ENSDART00000155362
|
si:dkeyp-94h10.5
|
si:dkeyp-94h10.5 |
| chr1_+_52518176 | 0.37 |
ENSDART00000003278
|
tacr3l
|
tachykinin receptor 3-like |
| chr7_-_5316901 | 0.37 |
ENSDART00000181505
ENSDART00000124367 |
si:cabz01074946.1
|
si:cabz01074946.1 |
| chr2_+_34138376 | 0.37 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr9_-_56231387 | 0.37 |
ENSDART00000149851
|
rpl31
|
ribosomal protein L31 |
| chr3_+_62161184 | 0.37 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr6_-_54845083 | 0.37 |
ENSDART00000148815
|
csrp1b
|
cysteine and glycine-rich protein 1b |
| chr14_-_28568107 | 0.37 |
ENSDART00000042850
ENSDART00000145502 |
insb
|
preproinsulin b |
| chr21_+_45223194 | 0.36 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr20_-_5267600 | 0.35 |
ENSDART00000099258
|
cyp46a1.4
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 4 |
| chr12_+_28854410 | 0.35 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr8_-_23573084 | 0.35 |
ENSDART00000139084
|
wasb
|
Wiskott-Aldrich syndrome (eczema-thrombocytopenia) b |
| chr16_-_52821023 | 0.35 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr13_-_44738574 | 0.35 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr21_+_6961635 | 0.35 |
ENSDART00000137281
|
si:ch211-93g21.2
|
si:ch211-93g21.2 |
| chr16_-_40508624 | 0.34 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr7_-_27033080 | 0.33 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr1_-_13968153 | 0.32 |
ENSDART00000103383
|
elf2b
|
E74-like factor 2b (ets domain transcription factor) |
| chr18_-_14691727 | 0.32 |
ENSDART00000010129
|
pdf
|
peptide deformylase, mitochondrial |
| chr25_+_34845115 | 0.31 |
ENSDART00000061996
|
tmem231
|
transmembrane protein 231 |
| chr12_+_46960579 | 0.31 |
ENSDART00000149032
|
oat
|
ornithine aminotransferase |
| chr8_+_23213320 | 0.31 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr8_+_471342 | 0.30 |
ENSDART00000167205
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr2_+_22795494 | 0.30 |
ENSDART00000042255
|
rab6bb
|
RAB6B, member RAS oncogene family b |
| chr24_-_25186895 | 0.30 |
ENSDART00000081029
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr25_-_36286186 | 0.29 |
ENSDART00000182398
|
slc7a10b
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10b |
| chr5_+_37785152 | 0.29 |
ENSDART00000053511
ENSDART00000189812 |
myo1ca
|
myosin Ic, paralog a |
| chr11_+_3254252 | 0.28 |
ENSDART00000123568
|
pmela
|
premelanosome protein a |
| chr6_-_13200585 | 0.28 |
ENSDART00000185321
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr8_-_25011157 | 0.28 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr14_+_8476769 | 0.28 |
ENSDART00000131773
|
dnajc4
|
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr1_-_44940830 | 0.27 |
ENSDART00000097500
ENSDART00000134464 ENSDART00000137216 |
tmem176
|
transmembrane protein 176 |
| chr17_-_43428397 | 0.26 |
ENSDART00000043866
|
plk4
|
polo-like kinase 4 (Drosophila) |
| chr10_-_33379850 | 0.26 |
ENSDART00000186924
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr22_+_21972824 | 0.25 |
ENSDART00000183708
ENSDART00000133739 |
gna15.3
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 3 |
| chr13_+_28512863 | 0.25 |
ENSDART00000043117
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr13_+_45709380 | 0.25 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr3_-_2589704 | 0.25 |
ENSDART00000187239
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr23_+_36653376 | 0.25 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr2_+_47581997 | 0.25 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr17_-_53329704 | 0.24 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr21_-_2238277 | 0.24 |
ENSDART00000165754
|
si:dkey-50i6.6
|
si:dkey-50i6.6 |
| chr17_-_51938663 | 0.24 |
ENSDART00000179784
|
ERG28
|
ergosterol biosynthesis 28 homolog |
| chr19_+_34742706 | 0.24 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr3_+_26223376 | 0.24 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr13_-_32635859 | 0.23 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr17_+_24006792 | 0.23 |
ENSDART00000122415
|
si:ch211-63b16.4
|
si:ch211-63b16.4 |
| chr6_-_32314913 | 0.23 |
ENSDART00000051779
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr4_-_12725513 | 0.23 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr17_+_53284040 | 0.23 |
ENSDART00000165368
|
atxn3
|
ataxin 3 |
| chr19_+_30387999 | 0.22 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr21_+_21339311 | 0.22 |
ENSDART00000137693
ENSDART00000161820 |
sdhaf1
si:ch211-191j22.3
|
succinate dehydrogenase complex assembly factor 1 si:ch211-191j22.3 |
| chr6_+_14980761 | 0.22 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr23_+_43855618 | 0.21 |
ENSDART00000156842
|
cnga1
|
cyclic nucleotide gated channel alpha 1 |
| chr16_+_40508882 | 0.21 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr21_-_9991502 | 0.21 |
ENSDART00000169320
|
esm1
|
endothelial cell-specific molecule 1 |
| chr7_-_24644893 | 0.20 |
ENSDART00000048921
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr15_+_5973909 | 0.20 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr25_-_35996141 | 0.20 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr5_-_6508250 | 0.20 |
ENSDART00000060535
|
crybb3
|
crystallin, beta B3 |
| chr12_-_46959990 | 0.20 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr23_+_32499916 | 0.20 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr6_+_28205284 | 0.19 |
ENSDART00000160707
ENSDART00000190509 |
smx5
LSM2 (1 of many)
|
smx5 si:ch73-14h10.2 |
| chr7_-_38638809 | 0.19 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr8_-_30791266 | 0.19 |
ENSDART00000062220
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr6_-_39218609 | 0.19 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr1_-_7582859 | 0.19 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr20_-_26929685 | 0.18 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr1_+_20635190 | 0.18 |
ENSDART00000145418
ENSDART00000148518 ENSDART00000139461 ENSDART00000102969 ENSDART00000166479 |
spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr19_+_9533008 | 0.18 |
ENSDART00000104607
|
fam131ba
|
family with sequence similarity 131, member Ba |
| chr12_+_26632448 | 0.18 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr25_-_13676682 | 0.18 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr19_+_30388186 | 0.18 |
ENSDART00000103474
|
tspan13b
|
tetraspanin 13b |
| chr7_+_17374560 | 0.18 |
ENSDART00000172272
ENSDART00000073467 |
nitr7a
|
novel immune-type receptor 7a |
| chr21_+_5915041 | 0.18 |
ENSDART00000151370
|
prodha
|
proline dehydrogenase (oxidase) 1a |
| chr21_-_4244514 | 0.17 |
ENSDART00000065880
|
uck1
|
uridine-cytidine kinase 1 |
| chr25_+_25438322 | 0.17 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr7_-_23873173 | 0.17 |
ENSDART00000101408
|
zgc:162171
|
zgc:162171 |
| chr21_-_2248239 | 0.17 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr8_-_30791089 | 0.17 |
ENSDART00000147332
|
gstt1a
|
glutathione S-transferase theta 1a |
| chr8_+_23009662 | 0.17 |
ENSDART00000030885
|
uckl1a
|
uridine-cytidine kinase 1-like 1a |
| chr21_-_26483237 | 0.17 |
ENSDART00000169072
ENSDART00000147947 |
ankhd1
|
ankyrin repeat and KH domain containing 1 |
| chr1_+_24366110 | 0.16 |
ENSDART00000139913
|
pimr178
|
Pim proto-oncogene, serine/threonine kinase, related 178 |
| chr4_-_8093753 | 0.16 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr16_+_32131568 | 0.16 |
ENSDART00000171454
|
sphk2
|
sphingosine kinase 2 |
| chr17_-_9995667 | 0.16 |
ENSDART00000148463
|
snx6
|
sorting nexin 6 |
| chr24_-_12938922 | 0.16 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr22_-_13165186 | 0.16 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr16_-_19349318 | 0.15 |
ENSDART00000020821
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr3_+_57997980 | 0.15 |
ENSDART00000168477
ENSDART00000193840 |
pycr1a
|
pyrroline-5-carboxylate reductase 1a |
| chr20_+_26349002 | 0.14 |
ENSDART00000152842
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr25_+_7671640 | 0.14 |
ENSDART00000145367
|
kcnj11l
|
potassium inwardly-rectifying channel, subfamily J, member 11, like |
| chr22_-_30948457 | 0.14 |
ENSDART00000139887
|
ssuh2.1
|
ssu-2 homolog, tandem duplicate 1 |
| chr11_-_21014572 | 0.14 |
ENSDART00000042220
|
slc6a14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chr20_+_49119633 | 0.14 |
ENSDART00000151435
|
cd109
|
CD109 molecule |
| chr24_+_2961098 | 0.14 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr12_-_10275320 | 0.14 |
ENSDART00000170078
|
mpp2b
|
membrane protein, palmitoylated 2b (MAGUK p55 subfamily member 2) |
| chr11_-_41130239 | 0.14 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr8_+_44926946 | 0.13 |
ENSDART00000098567
|
zgc:154046
|
zgc:154046 |
| chr10_+_3875716 | 0.13 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr7_-_41916505 | 0.13 |
ENSDART00000185785
|
dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2 |
| chr22_-_28226948 | 0.13 |
ENSDART00000147686
|
si:dkey-222p3.1
|
si:dkey-222p3.1 |
| chr1_+_44941031 | 0.13 |
ENSDART00000141145
|
si:dkey-9i23.16
|
si:dkey-9i23.16 |
| chr16_+_34479064 | 0.13 |
ENSDART00000159965
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr25_-_3759322 | 0.12 |
ENSDART00000088077
|
zgc:158398
|
zgc:158398 |
| chr2_-_32386598 | 0.12 |
ENSDART00000145575
|
ubtfl
|
upstream binding transcription factor, like |
| chr2_+_47582681 | 0.12 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr11_-_3574311 | 0.12 |
ENSDART00000167064
|
si:dkey-33m11.7
|
si:dkey-33m11.7 |
| chr2_+_35854242 | 0.12 |
ENSDART00000134918
|
dhx9
|
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr5_+_31860043 | 0.12 |
ENSDART00000036235
ENSDART00000140541 |
iscub
|
iron-sulfur cluster assembly enzyme b |
| chr25_-_6557854 | 0.12 |
ENSDART00000181740
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr3_+_28831450 | 0.12 |
ENSDART00000055422
|
flr
|
fleer |
| chr8_+_68864 | 0.11 |
ENSDART00000164574
|
prr16
|
proline rich 16 |
| chr19_+_10832092 | 0.11 |
ENSDART00000191851
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr17_+_45404758 | 0.11 |
ENSDART00000147557
|
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr12_-_47899497 | 0.11 |
ENSDART00000162219
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr17_+_8323348 | 0.11 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr8_-_20858676 | 0.11 |
ENSDART00000018515
|
dohh
|
deoxyhypusine hydroxylase/monooxygenase |
| chr7_+_23875269 | 0.11 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr6_-_18698006 | 0.10 |
ENSDART00000170128
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr3_-_58798377 | 0.10 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr19_+_8973042 | 0.10 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr23_-_4404105 | 0.10 |
ENSDART00000146941
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr17_+_43468732 | 0.10 |
ENSDART00000055487
|
chmp3
|
charged multivesicular body protein 3 |
| chr22_+_24378064 | 0.10 |
ENSDART00000180592
|
BX649608.2
|
|
| chr19_+_34311374 | 0.10 |
ENSDART00000086617
|
gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr7_+_6879534 | 0.09 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr6_+_12326267 | 0.09 |
ENSDART00000155101
|
si:dkey-276j7.3
|
si:dkey-276j7.3 |
| chr19_+_5480327 | 0.09 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr7_+_4384863 | 0.08 |
ENSDART00000042955
ENSDART00000134653 |
slc12a10.3
|
slc12a10.3 solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 3 |
| chr3_-_32611833 | 0.08 |
ENSDART00000151282
|
si:ch73-248e21.5
|
si:ch73-248e21.5 |
| chr20_-_17259573 | 0.08 |
ENSDART00000166397
|
AL928808.1
|
|
| chr12_-_32426685 | 0.08 |
ENSDART00000105574
|
enpp7.2
|
ectonucleotide pyrophosphatase/phosphodiesterase 7, tandem duplicate 2 |
| chr13_+_4205724 | 0.08 |
ENSDART00000134105
|
dlk2
|
delta-like 2 homolog (Drosophila) |
| chr9_-_23944470 | 0.08 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr6_-_18698669 | 0.08 |
ENSDART00000168554
ENSDART00000175205 |
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr3_+_25849560 | 0.08 |
ENSDART00000007119
|
mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr16_+_24681177 | 0.07 |
ENSDART00000058956
ENSDART00000189335 |
ywhabl
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide like |
| chr9_+_31282161 | 0.07 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr23_+_19606291 | 0.07 |
ENSDART00000139415
|
flnb
|
filamin B, beta (actin binding protein 278) |
| chr14_-_25985698 | 0.07 |
ENSDART00000172909
ENSDART00000123053 |
atox1
|
antioxidant 1 copper chaperone |
| chr13_-_40726865 | 0.07 |
ENSDART00000099847
|
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr7_+_21272833 | 0.07 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr21_-_4032650 | 0.07 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr6_-_18698456 | 0.07 |
ENSDART00000166587
|
rhbdl3
|
rhomboid, veinlet-like 3 (Drosophila) |
| chr7_-_18656069 | 0.06 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr21_-_31238244 | 0.06 |
ENSDART00000159678
|
tpst1l
|
tyrosylprotein sulfotransferase 1, like |
| chr3_+_38793499 | 0.06 |
ENSDART00000083394
|
kcnj19a
|
potassium voltage-gated channel subfamily J member 19a |
| chr11_+_44579865 | 0.06 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr7_+_50464500 | 0.06 |
ENSDART00000191356
|
serinc4
|
serine incorporator 4 |
| chr3_+_32689707 | 0.06 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr7_-_17779644 | 0.06 |
ENSDART00000128504
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr2_+_47582488 | 0.06 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr2_-_6039757 | 0.06 |
ENSDART00000013079
|
scp2a
|
sterol carrier protein 2a |
| chr5_+_69868911 | 0.06 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr22_+_17192767 | 0.06 |
ENSDART00000130810
ENSDART00000183006 |
atpaf1
|
ATP synthase mitochondrial F1 complex assembly factor 1 |
| chr21_-_40083432 | 0.06 |
ENSDART00000141160
ENSDART00000191195 |
slc13a5a
|
info solute carrier family 13 (sodium-dependent citrate transporter), member 5a |
| chr6_-_43007264 | 0.06 |
ENSDART00000124623
|
si:ch73-361p23.3
|
si:ch73-361p23.3 |
| chr24_+_14713776 | 0.06 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr13_-_15865335 | 0.06 |
ENSDART00000135186
|
si:ch211-57f7.7
|
si:ch211-57f7.7 |
| chr12_+_20412564 | 0.05 |
ENSDART00000186783
|
arhgap17a
|
Rho GTPase activating protein 17a |
| chr5_-_24270989 | 0.05 |
ENSDART00000146251
|
si:ch211-137i24.12
|
si:ch211-137i24.12 |
| chr2_+_30960351 | 0.05 |
ENSDART00000141575
|
lpin2
|
lipin 2 |
| chr18_-_34156765 | 0.05 |
ENSDART00000059400
|
c18h3orf33
|
c18h3orf33 homolog (H. sapiens) |
| chr14_-_1280907 | 0.05 |
ENSDART00000186150
|
CABZ01081490.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of irx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 0.4 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.6 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.3 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.3 | GO:0042940 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.1 | 0.3 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.1 | 1.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.4 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.3 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.3 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.7 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.4 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.5 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 0.9 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.4 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.0 | GO:0019474 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.4 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.2 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 0.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.7 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.5 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.5 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.2 | 0.5 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.7 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.2 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.7 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.2 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |