Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
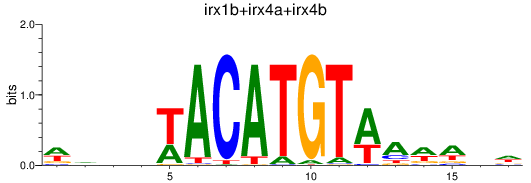
Results for irx1b+irx4a+irx4b
Z-value: 0.28
Transcription factors associated with irx1b+irx4a+irx4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irx4a
|
ENSDARG00000035648 | iroquois homeobox 4a |
|
irx4b
|
ENSDARG00000036051 | iroquois homeobox 4b |
|
irx1b
|
ENSDARG00000056594 | iroquois homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irx4b | dr11_v1_chr19_+_28115801_28115801 | 0.64 | 4.6e-03 | Click! |
| irx1b | dr11_v1_chr19_-_28130658_28130658 | -0.21 | 3.9e-01 | Click! |
| irx4a | dr11_v1_chr16_-_383664_383664 | -0.09 | 7.4e-01 | Click! |
Activity profile of irx1b+irx4a+irx4b motif
Sorted Z-values of irx1b+irx4a+irx4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_2908764 | 0.75 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr10_+_45345574 | 0.56 |
ENSDART00000166085
|
ppiab
|
peptidylprolyl isomerase Ab (cyclophilin A) |
| chr8_-_23783633 | 0.44 |
ENSDART00000132657
|
si:ch211-163l21.7
|
si:ch211-163l21.7 |
| chr5_+_34578479 | 0.43 |
ENSDART00000016314
|
gfm2
|
G elongation factor, mitochondrial 2 |
| chr19_+_3653976 | 0.42 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr20_+_23440632 | 0.40 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr20_+_30939178 | 0.37 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr1_+_53919110 | 0.35 |
ENSDART00000020680
|
nup133
|
nucleoporin 133 |
| chr8_-_50147948 | 0.29 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr14_-_10617127 | 0.25 |
ENSDART00000154299
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr10_+_8629275 | 0.25 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr7_+_73332486 | 0.24 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr12_-_34035364 | 0.23 |
ENSDART00000087065
|
timp2a
|
TIMP metallopeptidase inhibitor 2a |
| chr18_+_5549672 | 0.19 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr17_-_14705039 | 0.19 |
ENSDART00000154281
ENSDART00000123550 |
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr16_-_44709832 | 0.17 |
ENSDART00000156784
|
si:ch211-151m7.6
|
si:ch211-151m7.6 |
| chr7_-_44672095 | 0.14 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr21_+_43882274 | 0.13 |
ENSDART00000075672
|
sra1
|
steroid receptor RNA activator 1 |
| chr7_-_13362590 | 0.13 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr20_-_23440955 | 0.11 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr13_-_43149063 | 0.11 |
ENSDART00000099601
|
vsir
|
V-set immunoregulatory receptor |
| chr2_-_13333932 | 0.10 |
ENSDART00000150238
ENSDART00000168258 |
si:dkey-185p13.1
vps4b
|
si:dkey-185p13.1 vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr23_+_19701587 | 0.10 |
ENSDART00000104425
|
dnase1l1
|
deoxyribonuclease I-like 1 |
| chr15_+_33991928 | 0.10 |
ENSDART00000170177
|
vwde
|
von Willebrand factor D and EGF domains |
| chr10_-_45002196 | 0.09 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr16_-_49781481 | 0.09 |
ENSDART00000127513
|
znf385d
|
zinc finger protein 385D |
| chr22_+_16320076 | 0.08 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr7_-_31830936 | 0.08 |
ENSDART00000052514
ENSDART00000129720 |
cars
|
cysteinyl-tRNA synthetase |
| chr7_+_56577906 | 0.07 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr15_-_41734639 | 0.07 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr16_-_45398408 | 0.07 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr13_+_23677949 | 0.06 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr15_-_14375452 | 0.06 |
ENSDART00000160675
ENSDART00000164028 ENSDART00000171642 |
dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr15_+_37331585 | 0.05 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr8_+_29986265 | 0.05 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr2_-_31521764 | 0.04 |
ENSDART00000140523
|
mrc1b
|
mannose receptor, C type 1b |
| chr23_+_40275601 | 0.03 |
ENSDART00000076876
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr15_+_11381532 | 0.03 |
ENSDART00000124172
|
si:ch73-321d9.2
|
si:ch73-321d9.2 |
| chr23_+_40275400 | 0.02 |
ENSDART00000184259
|
fam46ab
|
family with sequence similarity 46, member Ab |
| chr14_+_11762991 | 0.01 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr3_-_34100700 | 0.01 |
ENSDART00000151628
|
ighv6-1
|
immunoglobulin heavy variable 6-1 |
| chr1_-_7951002 | 0.01 |
ENSDART00000138187
|
si:dkey-79f11.8
|
si:dkey-79f11.8 |
| chr1_+_961607 | 0.01 |
ENSDART00000184660
|
n6amt1
|
N-6 adenine-specific DNA methyltransferase 1 |
| chr4_-_5691257 | 0.00 |
ENSDART00000110497
|
tmem63a
|
transmembrane protein 63A |
| chr11_+_14284866 | 0.00 |
ENSDART00000163729
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irx1b+irx4a+irx4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 0.4 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.4 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0097032 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.2 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.4 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0097108 | hedgehog family protein binding(GO:0097108) |