Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
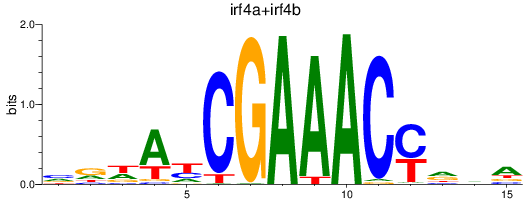
Results for irf4a+irf4b
Z-value: 1.05
Transcription factors associated with irf4a+irf4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf4a
|
ENSDARG00000006560 | interferon regulatory factor 4a |
|
irf4b
|
ENSDARG00000055374 | interferon regulatory factor 4b |
|
irf4b
|
ENSDARG00000115381 | interferon regulatory factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf4a | dr11_v1_chr2_-_879800_879800 | 0.89 | 5.4e-07 | Click! |
| irf4b | dr11_v1_chr20_+_26556174_26556174 | 0.71 | 8.9e-04 | Click! |
Activity profile of irf4a+irf4b motif
Sorted Z-values of irf4a+irf4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_27242498 | 2.74 |
ENSDART00000152609
ENSDART00000152170 |
si:dkey-11c5.11
|
si:dkey-11c5.11 |
| chr14_-_30960470 | 2.72 |
ENSDART00000129989
|
si:ch211-191o15.6
|
si:ch211-191o15.6 |
| chr19_-_7110617 | 2.45 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr14_+_33329761 | 2.22 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr4_-_12795030 | 2.06 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr5_+_65157576 | 1.80 |
ENSDART00000159599
|
cfap157
|
cilia and flagella associated protein 157 |
| chr4_-_8611841 | 1.80 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr6_+_15762647 | 1.79 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr4_-_12795436 | 1.71 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr14_+_33329420 | 1.67 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr19_-_6303099 | 1.67 |
ENSDART00000133371
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr10_+_5060191 | 1.61 |
ENSDART00000145908
ENSDART00000122397 |
carm1l
|
coactivator-associated arginine methyltransferase 1, like |
| chr25_+_16646113 | 1.60 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr13_+_17694845 | 1.44 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr20_-_3911546 | 1.41 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr16_+_29492937 | 1.40 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr1_-_7570181 | 1.40 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr25_+_36045072 | 1.25 |
ENSDART00000126326
|
rpgrip1l
|
RPGRIP1-like |
| chr8_-_27687095 | 1.21 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr18_+_17624295 | 1.20 |
ENSDART00000151865
ENSDART00000190408 ENSDART00000189204 ENSDART00000188894 ENSDART00000033762 ENSDART00000193471 |
nlrc5
|
NLR family, CARD domain containing 5 |
| chr15_-_17071328 | 1.17 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr4_-_815871 | 1.16 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr4_-_17669881 | 1.15 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr9_-_32837860 | 1.11 |
ENSDART00000142227
|
mxe
|
myxovirus (influenza virus) resistance E |
| chr9_-_30576522 | 1.10 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr10_-_5821584 | 1.09 |
ENSDART00000166388
|
il6st
|
interleukin 6 signal transducer |
| chr4_-_26107841 | 1.05 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr6_+_11249706 | 1.02 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr11_-_42472941 | 0.95 |
ENSDART00000166624
|
arf4b
|
ADP-ribosylation factor 4b |
| chr1_-_7582859 | 0.94 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr5_+_13385837 | 0.94 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr17_+_43895158 | 0.93 |
ENSDART00000111123
|
msh4
|
mutS homolog 4 |
| chr2_-_24554416 | 0.92 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr6_+_11250033 | 0.87 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr15_+_2421432 | 0.86 |
ENSDART00000193772
|
hephl1a
|
hephaestin-like 1a |
| chr6_-_18400548 | 0.85 |
ENSDART00000179797
ENSDART00000164891 |
trim25
|
tripartite motif containing 25 |
| chr17_+_35362851 | 0.84 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr13_-_50200348 | 0.82 |
ENSDART00000038391
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr2_-_26620803 | 0.76 |
ENSDART00000003946
ENSDART00000126826 |
tmem59
|
transmembrane protein 59 |
| chr1_-_35924495 | 0.76 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr25_-_774350 | 0.74 |
ENSDART00000166321
ENSDART00000160386 |
IRAK4
|
interleukin 1 receptor associated kinase 4 |
| chr19_+_10832092 | 0.72 |
ENSDART00000191851
|
tomm40l
|
translocase of outer mitochondrial membrane 40 homolog, like |
| chr18_-_23875370 | 0.67 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_-_18229169 | 0.66 |
ENSDART00000131764
ENSDART00000143036 ENSDART00000145986 |
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr13_-_45475289 | 0.66 |
ENSDART00000043345
|
rsrp1
|
arginine/serine-rich protein 1 |
| chr13_-_50200042 | 0.65 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr7_-_19614916 | 0.64 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr5_+_69716458 | 0.63 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr14_-_25309058 | 0.62 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr21_-_22543611 | 0.61 |
ENSDART00000177084
|
myo5b
|
myosin VB |
| chr2_+_37975026 | 0.61 |
ENSDART00000034802
|
si:rp71-1g18.13
|
si:rp71-1g18.13 |
| chr6_+_39493864 | 0.61 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr9_+_51147764 | 0.61 |
ENSDART00000188480
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr2_+_2818645 | 0.60 |
ENSDART00000163587
|
rock1
|
Rho-associated, coiled-coil containing protein kinase 1 |
| chr7_+_19615056 | 0.60 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr20_+_38525567 | 0.56 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr21_+_45626136 | 0.54 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr11_-_6206520 | 0.53 |
ENSDART00000150199
ENSDART00000148246 ENSDART00000019440 |
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr4_-_6373735 | 0.52 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr15_-_41734639 | 0.51 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr8_+_50983551 | 0.49 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr22_-_17474583 | 0.49 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr22_+_19365220 | 0.47 |
ENSDART00000132781
ENSDART00000135672 ENSDART00000153630 |
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr15_-_21702317 | 0.45 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr24_-_3407507 | 0.45 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr15_-_40175894 | 0.43 |
ENSDART00000156632
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr12_-_35936329 | 0.43 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr5_+_59397739 | 0.41 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr15_-_42760110 | 0.41 |
ENSDART00000152490
|
si:ch211-181d7.3
|
si:ch211-181d7.3 |
| chr15_+_2421729 | 0.38 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr9_+_51147380 | 0.38 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr17_-_23603263 | 0.38 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr6_-_1566581 | 0.36 |
ENSDART00000192993
|
trim107
|
tripartite motif containing 107 |
| chr14_-_25309360 | 0.34 |
ENSDART00000088940
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr1_+_31674297 | 0.32 |
ENSDART00000044214
|
wbp1lb
|
WW domain binding protein 1-like b |
| chr15_-_41749364 | 0.32 |
ENSDART00000155464
|
ftr73
|
finTRIM family, member 73 |
| chr22_-_17474781 | 0.32 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr7_-_8881514 | 0.31 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr3_-_29941357 | 0.30 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr4_+_11464255 | 0.29 |
ENSDART00000008584
|
gdi2
|
GDP dissociation inhibitor 2 |
| chr1_-_1885516 | 0.29 |
ENSDART00000122187
ENSDART00000131675 |
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr24_-_11467549 | 0.27 |
ENSDART00000082264
|
pxdc1b
|
PX domain containing 1b |
| chr8_-_51340931 | 0.27 |
ENSDART00000178209
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr4_-_18434924 | 0.25 |
ENSDART00000190271
|
socs2
|
suppressor of cytokine signaling 2 |
| chr20_-_35012093 | 0.25 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr18_-_16179129 | 0.25 |
ENSDART00000125353
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr20_-_51547464 | 0.22 |
ENSDART00000099486
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr20_-_25436389 | 0.22 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr22_-_10604526 | 0.22 |
ENSDART00000091777
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr21_-_38031038 | 0.22 |
ENSDART00000179483
ENSDART00000076238 |
rbm41
|
RNA binding motif protein 41 |
| chr20_+_2227860 | 0.22 |
ENSDART00000152321
|
tmem200a
|
transmembrane protein 200A |
| chr21_+_28728030 | 0.22 |
ENSDART00000097307
|
puraa
|
purine-rich element binding protein Aa |
| chr25_-_483808 | 0.21 |
ENSDART00000104720
|
si:ch1073-385f13.3
|
si:ch1073-385f13.3 |
| chr23_-_30045661 | 0.19 |
ENSDART00000122239
ENSDART00000103480 |
ccdc187
|
coiled-coil domain containing 187 |
| chr5_+_35502290 | 0.18 |
ENSDART00000191222
|
CR762390.1
|
|
| chr10_+_8101729 | 0.17 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr8_-_51340773 | 0.17 |
ENSDART00000060633
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr6_+_11250316 | 0.16 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr6_+_34512313 | 0.15 |
ENSDART00000102554
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr13_+_22476742 | 0.15 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr4_+_68562464 | 0.14 |
ENSDART00000192954
|
BX548011.4
|
|
| chr16_+_48753664 | 0.14 |
ENSDART00000155148
|
si:ch73-31d8.2
|
si:ch73-31d8.2 |
| chr19_-_791016 | 0.12 |
ENSDART00000037515
|
msto1
|
misato 1, mitochondrial distribution and morphology regulator |
| chr8_+_45361775 | 0.11 |
ENSDART00000015193
|
chmp4bb
|
charged multivesicular body protein 4Bb |
| chr14_+_25817246 | 0.11 |
ENSDART00000136733
|
glra1
|
glycine receptor, alpha 1 |
| chr21_+_25793970 | 0.09 |
ENSDART00000101217
|
CLDN4 (1 of many)
|
zgc:136892 |
| chr3_-_4501026 | 0.08 |
ENSDART00000163052
|
zgc:162198
|
zgc:162198 |
| chr5_-_45894802 | 0.08 |
ENSDART00000097648
|
crfb6
|
cytokine receptor family member b6 |
| chr18_-_23874929 | 0.08 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr18_-_23875219 | 0.07 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_+_47897734 | 0.07 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr18_-_24988645 | 0.07 |
ENSDART00000136434
ENSDART00000085735 |
chd2
|
chromodomain helicase DNA binding protein 2 |
| chr17_+_40879331 | 0.06 |
ENSDART00000046003
|
slc4a1ap
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr14_-_33585809 | 0.04 |
ENSDART00000023540
|
sash3
|
SAM and SH3 domain containing 3 |
| chr9_+_42269059 | 0.03 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr23_-_29553691 | 0.02 |
ENSDART00000053804
|
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr22_-_10605045 | 0.02 |
ENSDART00000184812
|
bap1
|
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr6_+_41186320 | 0.01 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr12_-_23658888 | 0.01 |
ENSDART00000088319
|
map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr17_-_23727978 | 0.01 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf4a+irf4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.0 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.3 | 0.9 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.3 | 0.8 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 0.8 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.3 | 1.8 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.2 | 1.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 2.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.2 | 0.6 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.2 | 3.4 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 1.0 | GO:0032728 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.2 | 1.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 0.6 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.4 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.1 | 1.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.5 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.9 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 1.2 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 1.0 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.1 | 1.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 2.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.2 | GO:0035521 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.9 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.6 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 1.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.8 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 1.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.9 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.2 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 1.4 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.0 | 1.1 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.8 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 3.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 0.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.6 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 2.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 0.7 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.6 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.2 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.4 | 1.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 0.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 1.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 2.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.1 | 1.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.6 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.9 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.7 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 1.0 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 5.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 1.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.8 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 1.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.8 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.8 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.6 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |