Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
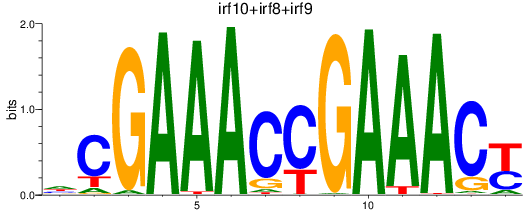
Results for irf10+irf8+irf9
Z-value: 0.82
Transcription factors associated with irf10+irf8+irf9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
irf9
|
ENSDARG00000016457 | interferon regulatory factor 9 |
|
irf10
|
ENSDARG00000027658 | interferon regulatory factor 10 |
|
irf8
|
ENSDARG00000056407 | interferon regulatory factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| irf9 | dr11_v1_chr12_+_13282797_13282807 | 0.95 | 1.1e-09 | Click! |
| irf10 | dr11_v1_chr23_+_642001_642052 | 0.91 | 1.5e-07 | Click! |
| irf8 | dr11_v1_chr18_+_30567945_30567945 | 0.62 | 6.3e-03 | Click! |
Activity profile of irf10+irf8+irf9 motif
Sorted Z-values of irf10+irf8+irf9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_59029485 | 2.59 |
ENSDART00000050140
|
CABZ01088367.1
|
|
| chr2_+_52122663 | 2.52 |
ENSDART00000111266
|
theg
|
theg spermatid protein |
| chr4_-_13567387 | 2.26 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr10_-_34889053 | 2.25 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr13_-_37608441 | 2.24 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr24_+_37902687 | 2.17 |
ENSDART00000042273
|
ccdc78
|
coiled-coil domain containing 78 |
| chr19_-_7115229 | 2.02 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr8_-_50525360 | 1.91 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr10_+_5060191 | 1.85 |
ENSDART00000145908
ENSDART00000122397 |
carm1l
|
coactivator-associated arginine methyltransferase 1, like |
| chr12_-_30583668 | 1.84 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr20_-_16223245 | 1.66 |
ENSDART00000014721
|
echdc2
|
enoyl CoA hydratase domain containing 2 |
| chr3_+_29941777 | 1.58 |
ENSDART00000113889
|
ifi35
|
interferon-induced protein 35 |
| chr2_-_24554416 | 1.57 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr15_-_17071328 | 1.56 |
ENSDART00000122617
|
si:ch211-24o10.6
|
si:ch211-24o10.6 |
| chr20_+_54356540 | 1.52 |
ENSDART00000143591
|
znf410
|
zinc finger protein 410 |
| chr6_+_29217392 | 1.52 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr19_+_7115223 | 1.47 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr4_-_12795030 | 1.47 |
ENSDART00000150427
|
b2m
|
beta-2-microglobulin |
| chr20_+_54356272 | 1.45 |
ENSDART00000145735
|
znf410
|
zinc finger protein 410 |
| chr10_-_9410068 | 1.42 |
ENSDART00000041382
|
morn5
|
MORN repeat containing 5 |
| chr9_-_30576522 | 1.38 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr19_-_7110617 | 1.25 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr15_-_31067589 | 1.21 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr18_+_26422124 | 1.20 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr1_-_47161996 | 1.18 |
ENSDART00000053153
|
mhc1zba
|
major histocompatibility complex class I ZBA |
| chr16_+_29492937 | 1.17 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr18_+_34667486 | 1.08 |
ENSDART00000163583
|
CR626874.1
|
|
| chr10_-_4375190 | 1.07 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr10_+_26645953 | 1.04 |
ENSDART00000131482
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr4_-_779796 | 1.03 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr13_-_37642890 | 1.02 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr13_+_27232848 | 0.98 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr25_-_27665978 | 0.97 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr4_-_26108053 | 0.93 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr15_+_2421432 | 0.93 |
ENSDART00000193772
|
hephl1a
|
hephaestin-like 1a |
| chr1_+_2129164 | 0.91 |
ENSDART00000074923
ENSDART00000124534 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr24_+_12945803 | 0.89 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr15_+_13984879 | 0.89 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr16_+_29514473 | 0.88 |
ENSDART00000034102
|
ctss2.2
|
cathepsin S, ortholog 2, tandem duplicate 2 |
| chr22_-_10580194 | 0.86 |
ENSDART00000105848
|
SHISA4 (1 of many)
|
si:dkey-42i9.7 |
| chr19_-_325584 | 0.85 |
ENSDART00000134266
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr3_-_23596532 | 0.85 |
ENSDART00000124921
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr15_-_21702317 | 0.84 |
ENSDART00000155824
|
IL4I1
|
si:dkey-40g16.6 |
| chr15_-_29162193 | 0.84 |
ENSDART00000138449
ENSDART00000099885 |
xaf1
|
XIAP associated factor 1 |
| chr21_+_8341774 | 0.83 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr15_-_47929455 | 0.81 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr22_+_12798569 | 0.81 |
ENSDART00000005720
|
stat1a
|
signal transducer and activator of transcription 1a |
| chr3_-_12026741 | 0.81 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr10_+_26646104 | 0.80 |
ENSDART00000187685
|
adgrg4b
|
adhesion G protein-coupled receptor G4b |
| chr22_+_38301365 | 0.79 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr13_+_28495419 | 0.79 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr23_+_642001 | 0.78 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr4_-_26107841 | 0.78 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr25_+_14092871 | 0.76 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr17_+_35362851 | 0.75 |
ENSDART00000137659
|
cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr25_+_4979446 | 0.74 |
ENSDART00000154131
ENSDART00000155537 |
si:ch73-265h17.1
|
si:ch73-265h17.1 |
| chr18_-_12270845 | 0.73 |
ENSDART00000143923
ENSDART00000125555 ENSDART00000062360 ENSDART00000062216 |
nup205
|
nucleoporin 205 |
| chr19_+_7124337 | 0.72 |
ENSDART00000031380
|
tap2a
|
transporter associated with antigen processing, subunit type a |
| chr20_+_38525567 | 0.72 |
ENSDART00000147787
|
znf512
|
zinc finger protein 512 |
| chr5_+_13373593 | 0.72 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr4_-_17669881 | 0.71 |
ENSDART00000066997
|
dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr13_-_45155792 | 0.70 |
ENSDART00000163556
|
runx3
|
runt-related transcription factor 3 |
| chr7_+_25053331 | 0.70 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr5_-_24869213 | 0.68 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr23_-_31974060 | 0.67 |
ENSDART00000168087
|
BX927210.1
|
|
| chr22_-_17499513 | 0.65 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr25_+_35942867 | 0.63 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr8_+_20679759 | 0.63 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr25_-_35524166 | 0.63 |
ENSDART00000156782
|
slc5a12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr11_-_23458792 | 0.60 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr18_+_16246806 | 0.60 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr22_+_1683185 | 0.60 |
ENSDART00000158109
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr23_+_642395 | 0.58 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr15_+_17321218 | 0.58 |
ENSDART00000143796
|
cltcb
|
clathrin, heavy chain b (Hc) |
| chr18_+_10884996 | 0.57 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr8_+_1187928 | 0.56 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr15_+_24588963 | 0.55 |
ENSDART00000155075
|
zgc:198241
|
zgc:198241 |
| chr19_-_18855513 | 0.54 |
ENSDART00000162708
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr1_+_2128970 | 0.54 |
ENSDART00000180074
ENSDART00000022019 ENSDART00000098059 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr12_-_4598237 | 0.54 |
ENSDART00000152489
|
irf3
|
interferon regulatory factor 3 |
| chr11_-_19775182 | 0.53 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr9_+_51147764 | 0.51 |
ENSDART00000188480
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr22_-_36750589 | 0.51 |
ENSDART00000010824
|
acy1
|
aminoacylase 1 |
| chr23_-_4409668 | 0.51 |
ENSDART00000081823
|
si:ch73-142c19.1
|
si:ch73-142c19.1 |
| chr12_+_27114199 | 0.51 |
ENSDART00000150017
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr7_-_66877058 | 0.50 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr12_+_38556462 | 0.50 |
ENSDART00000021069
ENSDART00000145377 |
rpl38
|
ribosomal protein L38 |
| chr12_-_290413 | 0.49 |
ENSDART00000152496
|
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr10_+_2899108 | 0.49 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr15_-_43873005 | 0.49 |
ENSDART00000190326
|
NOX4
|
NADPH oxidase 4 |
| chr5_+_8919698 | 0.48 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr3_+_1942219 | 0.48 |
ENSDART00000114520
|
zgc:165583
|
zgc:165583 |
| chr19_+_22085925 | 0.48 |
ENSDART00000185636
|
atp9b
|
ATPase phospholipid transporting 9B |
| chr22_-_17474583 | 0.48 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr13_-_50200042 | 0.47 |
ENSDART00000074230
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr8_-_27656765 | 0.47 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr2_-_43135700 | 0.47 |
ENSDART00000098284
|
ftr14
|
finTRIM family, member 14 |
| chr25_-_6223567 | 0.47 |
ENSDART00000067512
|
psma4
|
proteasome subunit alpha 4 |
| chr24_-_28419444 | 0.47 |
ENSDART00000105749
|
nrros
|
negative regulator of reactive oxygen species |
| chr21_+_27340682 | 0.47 |
ENSDART00000011305
|
dpp3
|
dipeptidyl-peptidase 3 |
| chr13_-_50200348 | 0.46 |
ENSDART00000038391
|
pkz
|
protein kinase containing Z-DNA binding domains |
| chr25_-_6420800 | 0.46 |
ENSDART00000153768
|
ptpn9a
|
protein tyrosine phosphatase, non-receptor type 9, a |
| chr17_+_23942268 | 0.45 |
ENSDART00000154153
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr16_-_9802449 | 0.45 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr15_+_2421729 | 0.45 |
ENSDART00000082294
ENSDART00000156428 |
hephl1a
|
hephaestin-like 1a |
| chr6_+_52947186 | 0.45 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr5_+_68267908 | 0.45 |
ENSDART00000130835
|
tnfsf13
|
TNF superfamily member 13 |
| chr15_-_42760110 | 0.45 |
ENSDART00000152490
|
si:ch211-181d7.3
|
si:ch211-181d7.3 |
| chr12_-_35936329 | 0.44 |
ENSDART00000166634
|
rnf213b
|
ring finger protein 213b |
| chr3_-_23596809 | 0.44 |
ENSDART00000156897
|
ube2z
|
ubiquitin-conjugating enzyme E2Z |
| chr16_-_9802998 | 0.44 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr7_-_19614916 | 0.44 |
ENSDART00000169029
|
zgc:194655
|
zgc:194655 |
| chr16_-_43344859 | 0.44 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr13_-_37631092 | 0.44 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr15_+_9861973 | 0.43 |
ENSDART00000170945
|
si:dkey-13m3.2
|
si:dkey-13m3.2 |
| chr12_-_290782 | 0.41 |
ENSDART00000152527
ENSDART00000105694 |
adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr2_+_45479841 | 0.40 |
ENSDART00000151856
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr7_+_19615056 | 0.40 |
ENSDART00000124752
ENSDART00000190297 |
si:ch211-212k18.15
|
si:ch211-212k18.15 |
| chr13_+_27232694 | 0.40 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr21_+_33503835 | 0.39 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr13_-_7474324 | 0.39 |
ENSDART00000162550
|
si:dkey-196h17.9
|
si:dkey-196h17.9 |
| chr19_-_18855717 | 0.38 |
ENSDART00000158192
|
ppt2
|
palmitoyl-protein thioesterase 2 |
| chr4_+_5842433 | 0.38 |
ENSDART00000124085
ENSDART00000179848 |
usp18
|
ubiquitin specific peptidase 18 |
| chr4_+_34417403 | 0.37 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr22_-_5958066 | 0.37 |
ENSDART00000145821
|
si:rp71-36a1.3
|
si:rp71-36a1.3 |
| chr1_-_50710468 | 0.37 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr3_-_61203203 | 0.37 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr7_+_15618127 | 0.37 |
ENSDART00000080160
|
lrrc28
|
leucine rich repeat containing 28 |
| chr23_-_37835794 | 0.37 |
ENSDART00000137358
|
cd40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr7_-_37563883 | 0.37 |
ENSDART00000148805
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr5_-_52784152 | 0.37 |
ENSDART00000169307
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr14_+_6159356 | 0.37 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr25_-_34979548 | 0.36 |
ENSDART00000153904
|
si:dkey-108k21.7
|
si:dkey-108k21.7 |
| chr16_-_31686602 | 0.36 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr22_+_9060699 | 0.36 |
ENSDART00000133993
|
BX248395.1
|
|
| chr3_-_45361573 | 0.35 |
ENSDART00000154796
|
il21r.1
|
interleukin 21 receptor, tandem duplicate 1 |
| chr10_+_3875716 | 0.35 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr17_+_24006792 | 0.35 |
ENSDART00000122415
|
si:ch211-63b16.4
|
si:ch211-63b16.4 |
| chr25_-_25550938 | 0.35 |
ENSDART00000150412
ENSDART00000103622 |
irf7
|
interferon regulatory factor 7 |
| chr6_+_52947699 | 0.35 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr7_-_5191797 | 0.35 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr23_-_37575030 | 0.35 |
ENSDART00000031875
|
tor1l3
|
torsin family 1 like 3 |
| chr9_+_51147380 | 0.34 |
ENSDART00000003913
|
ifih1
|
interferon induced with helicase C domain 1 |
| chr18_+_33580391 | 0.33 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr24_+_10310577 | 0.32 |
ENSDART00000141718
|
otulina
|
OTU deubiquitinase with linear linkage specificity a |
| chr22_-_17474781 | 0.32 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr9_-_40935934 | 0.32 |
ENSDART00000155604
|
C2orf88
|
Small membrane A-kinase anchor protein |
| chr2_+_30144979 | 0.31 |
ENSDART00000129365
ENSDART00000130344 |
kcnb2
|
potassium voltage-gated channel, Shab-related subfamily, member 2 |
| chr5_-_37820878 | 0.31 |
ENSDART00000165513
|
AL935065.1
|
Danio rerio uncharacterized LOC567472 (LOC567472), mRNA. |
| chr10_+_40623414 | 0.30 |
ENSDART00000187616
|
zgc:172131
|
zgc:172131 |
| chr22_-_17459587 | 0.29 |
ENSDART00000142267
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr22_+_8772363 | 0.29 |
ENSDART00000161461
|
si:dkey-182g1.10
|
si:dkey-182g1.10 |
| chr12_-_19151708 | 0.29 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr4_+_3358383 | 0.29 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr15_-_31043183 | 0.29 |
ENSDART00000100145
|
lgals9l1
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 1 |
| chr5_-_67661102 | 0.29 |
ENSDART00000013605
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_32834007 | 0.29 |
ENSDART00000157353
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr11_-_36350421 | 0.28 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr1_-_54938137 | 0.28 |
ENSDART00000147212
|
crtac1a
|
cartilage acidic protein 1a |
| chr1_-_1885516 | 0.28 |
ENSDART00000122187
ENSDART00000131675 |
si:ch211-132g1.3
|
si:ch211-132g1.3 |
| chr3_-_52661242 | 0.28 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr7_-_804515 | 0.28 |
ENSDART00000159359
|
rela
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr20_-_20402500 | 0.28 |
ENSDART00000190747
ENSDART00000185065 |
prkchb
|
protein kinase C, eta, b |
| chr25_-_35960229 | 0.28 |
ENSDART00000073434
|
snx20
|
sorting nexin 20 |
| chr4_+_40953320 | 0.28 |
ENSDART00000151912
|
znf1136
|
zinc finger protein 1136 |
| chr2_-_47870649 | 0.27 |
ENSDART00000142854
|
ftr05
|
finTRIM family, member 5 |
| chr5_+_38662125 | 0.27 |
ENSDART00000136949
|
si:dkey-58f10.13
|
si:dkey-58f10.13 |
| chr10_+_17776981 | 0.27 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr18_+_19990412 | 0.26 |
ENSDART00000155054
ENSDART00000090310 |
pias1b
|
protein inhibitor of activated STAT, 1b |
| chr6_+_58569808 | 0.26 |
ENSDART00000154393
|
helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr8_-_25422186 | 0.26 |
ENSDART00000113492
ENSDART00000131736 |
kcnq2a
|
potassium voltage-gated channel, KQT-like subfamily, member 2a |
| chr2_+_41926707 | 0.26 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr22_+_1560991 | 0.26 |
ENSDART00000190639
|
AL954741.4
|
|
| chr15_-_36369743 | 0.25 |
ENSDART00000155942
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr18_+_29402623 | 0.25 |
ENSDART00000014703
|
mafa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog a (paralog a) |
| chr16_-_5115993 | 0.25 |
ENSDART00000138654
|
ttk
|
ttk protein kinase |
| chr3_-_4455951 | 0.25 |
ENSDART00000193908
ENSDART00000074077 |
trim35-3
|
tripartite motif containing 35-3 |
| chr16_+_13855039 | 0.25 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr1_-_52443379 | 0.24 |
ENSDART00000144676
|
si:ch211-217k17.11
|
si:ch211-217k17.11 |
| chr16_+_36906693 | 0.24 |
ENSDART00000160645
|
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr4_+_18707174 | 0.24 |
ENSDART00000142343
|
wnt7ba
|
wingless-type MMTV integration site family, member 7Ba |
| chr3_-_1938588 | 0.23 |
ENSDART00000013001
ENSDART00000186405 |
zgc:152753
|
zgc:152753 |
| chr16_+_25806400 | 0.23 |
ENSDART00000154652
|
irgq1
|
immunity-related GTPase family, q1 |
| chr16_+_48678655 | 0.23 |
ENSDART00000150156
|
tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_+_32834760 | 0.22 |
ENSDART00000121562
|
cyldl
|
cylindromatosis (turban tumor syndrome), like |
| chr5_+_45895613 | 0.22 |
ENSDART00000083937
|
polk
|
polymerase (DNA directed) kappa |
| chr1_-_30510839 | 0.21 |
ENSDART00000168189
ENSDART00000174868 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr1_-_49530615 | 0.21 |
ENSDART00000113315
|
si:ch211-281g13.5
|
si:ch211-281g13.5 |
| chr15_-_34845414 | 0.20 |
ENSDART00000009892
|
gabbr1a
|
gamma-aminobutyric acid (GABA) B receptor, 1a |
| chr1_-_25177086 | 0.20 |
ENSDART00000144711
ENSDART00000177225 |
tmem154
|
transmembrane protein 154 |
| chr15_+_32268790 | 0.20 |
ENSDART00000154457
|
fhdc4
|
FH2 domain containing 4 |
| chr10_+_26689262 | 0.20 |
ENSDART00000123195
ENSDART00000132946 |
tmtops3b
|
teleost multiple tissue opsin 3b |
| chr16_+_43344475 | 0.19 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr2_-_38141997 | 0.19 |
ENSDART00000143469
|
trim110
|
tripartite motif containing 110 |
| chr6_-_32834385 | 0.19 |
ENSDART00000129803
|
zc3h3
|
zinc finger CCCH-type containing 3 |
| chr22_-_4778018 | 0.18 |
ENSDART00000143968
|
si:ch73-256j6.5
|
si:ch73-256j6.5 |
| chr6_+_38427357 | 0.18 |
ENSDART00000148678
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_-_17028255 | 0.18 |
ENSDART00000152678
|
ifit10
|
interferon-induced protein with tetratricopeptide repeats 10 |
| chr4_+_43522680 | 0.17 |
ENSDART00000182252
|
si:dkeyp-53e4.4
|
si:dkeyp-53e4.4 |
| chr11_+_25481046 | 0.17 |
ENSDART00000065940
|
opn1lw2
|
opsin 1 (cone pigments), long-wave-sensitive, 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of irf10+irf8+irf9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 0.4 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.3 | 2.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 1.5 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.2 | 0.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 7.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.5 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.2 | 0.9 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.2 | 0.5 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 0.9 | GO:0032608 | interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.8 | GO:0021572 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.1 | 0.6 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.1 | 0.4 | GO:2000737 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) negative regulation of stem cell differentiation(GO:2000737) |
| 0.1 | 0.8 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 1.5 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.1 | 0.5 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.5 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.5 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.4 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.3 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 1.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 1.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.6 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.1 | 0.7 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.6 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 0.2 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.7 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.7 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 1.0 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 2.2 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 1.0 | GO:0034341 | response to interferon-gamma(GO:0034341) cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 0.8 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.8 | GO:0010952 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.2 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 0.3 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.5 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.6 | GO:0051607 | defense response to virus(GO:0051607) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.2 | 1.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 0.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.5 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.9 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.6 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.3 | 1.9 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.2 | 0.7 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.2 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.9 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.2 | 1.7 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 0.5 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.2 | 0.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 0.6 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.1 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.6 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 1.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 1.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.4 | GO:0015440 | peptide antigen-transporting ATPase activity(GO:0015433) peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) |
| 0.1 | 0.9 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.5 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.9 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.2 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 1.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.7 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 3.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.4 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.7 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |