Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
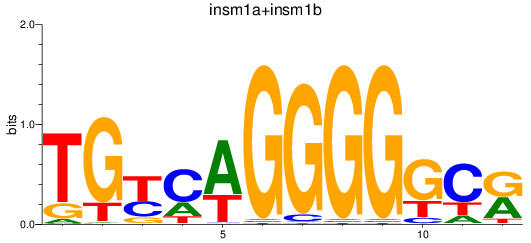
Results for insm1a+insm1b
Z-value: 0.48
Transcription factors associated with insm1a+insm1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
insm1b
|
ENSDARG00000053301 | insulinoma-associated 1b |
|
insm1a
|
ENSDARG00000091756 | insulinoma-associated 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| insm1b | dr11_v1_chr17_+_41463942_41463942 | -0.58 | 1.1e-02 | Click! |
| insm1a | dr11_v1_chr20_-_48485354_48485354 | -0.54 | 2.1e-02 | Click! |
Activity profile of insm1a+insm1b motif
Sorted Z-values of insm1a+insm1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_23697217 | 1.31 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr20_-_487783 | 1.27 |
ENSDART00000162218
|
col10a1b
|
collagen, type X, alpha 1b |
| chr3_-_40051425 | 1.08 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr7_+_67699009 | 1.04 |
ENSDART00000192810
|
zgc:162592
|
zgc:162592 |
| chr18_+_45573416 | 1.00 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr4_-_25908871 | 0.97 |
ENSDART00000066949
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr18_+_45573251 | 0.95 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr19_-_33996277 | 0.91 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr3_-_43646733 | 0.88 |
ENSDART00000180959
|
axin1
|
axin 1 |
| chr3_-_43650189 | 0.87 |
ENSDART00000161127
|
axin1
|
axin 1 |
| chr13_-_2981472 | 0.87 |
ENSDART00000184820
|
CABZ01087623.1
|
|
| chr8_-_18899427 | 0.86 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr17_+_1360192 | 0.86 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr12_-_24832297 | 0.86 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr5_-_4297459 | 0.84 |
ENSDART00000018895
|
srrt
|
serrate RNA effector molecule homolog (Arabidopsis) |
| chr18_-_127558 | 0.83 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_+_23826985 | 0.81 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr25_-_25575717 | 0.81 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr8_+_23827571 | 0.81 |
ENSDART00000040362
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr8_+_14959587 | 0.79 |
ENSDART00000138548
ENSDART00000041697 |
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr20_-_25644131 | 0.78 |
ENSDART00000138997
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr11_+_13041341 | 0.78 |
ENSDART00000166126
ENSDART00000162074 ENSDART00000170889 |
btf3l4
|
basic transcription factor 3-like 4 |
| chr1_-_40341306 | 0.68 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr17_+_6793001 | 0.67 |
ENSDART00000030773
|
foxo3a
|
forkhead box O3A |
| chr25_-_25575241 | 0.66 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr18_+_18000695 | 0.65 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr2_-_37098785 | 0.64 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr18_-_127873 | 0.63 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr2_+_15100742 | 0.63 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr2_+_7132292 | 0.63 |
ENSDART00000153404
ENSDART00000012119 |
zgc:110366
|
zgc:110366 |
| chr20_-_25643667 | 0.62 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr15_-_19128705 | 0.61 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr5_+_43006422 | 0.59 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr24_-_25166720 | 0.58 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr14_+_45350008 | 0.58 |
ENSDART00000025749
|
ttc9c
|
tetratricopeptide repeat domain 9C |
| chr9_+_30421489 | 0.56 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr14_-_6402769 | 0.55 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr5_-_37044262 | 0.54 |
ENSDART00000166670
|
si:dkeyp-110c7.8
|
si:dkeyp-110c7.8 |
| chr10_+_6383270 | 0.53 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr2_-_10564019 | 0.52 |
ENSDART00000132167
|
ccdc18
|
coiled-coil domain containing 18 |
| chr3_-_55328548 | 0.52 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr11_-_6877973 | 0.50 |
ENSDART00000160271
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr14_-_30983011 | 0.47 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr15_-_4616816 | 0.46 |
ENSDART00000160191
ENSDART00000161721 ENSDART00000144949 |
eif4h
|
eukaryotic translation initiation factor 4h |
| chr3_+_18437397 | 0.45 |
ENSDART00000136243
ENSDART00000184882 ENSDART00000135470 |
tbc1d16
|
TBC1 domain family, member 16 |
| chr20_+_2741802 | 0.45 |
ENSDART00000152135
|
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr8_-_1264893 | 0.44 |
ENSDART00000190371
|
cdc14b
|
cell division cycle 14B |
| chr7_-_30605089 | 0.43 |
ENSDART00000173775
ENSDART00000173789 ENSDART00000166046 ENSDART00000111626 |
rnf111
|
ring finger protein 111 |
| chr15_+_4988189 | 0.43 |
ENSDART00000142995
ENSDART00000062852 |
spcs2
|
signal peptidase complex subunit 2 |
| chr21_-_21526740 | 0.43 |
ENSDART00000142690
|
diablob
|
diablo, IAP-binding mitochondrial protein b |
| chr23_-_452365 | 0.41 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr18_+_18612388 | 0.41 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr24_-_28306484 | 0.39 |
ENSDART00000148618
|
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr13_-_36680531 | 0.39 |
ENSDART00000085298
|
l2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr25_+_32496723 | 0.38 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr13_-_49666615 | 0.38 |
ENSDART00000148083
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr23_+_19594608 | 0.38 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr18_+_17493859 | 0.37 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr25_+_32496877 | 0.37 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr23_-_27442544 | 0.36 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr19_+_9032073 | 0.36 |
ENSDART00000127755
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr11_-_11878099 | 0.35 |
ENSDART00000188314
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr20_-_34663209 | 0.34 |
ENSDART00000132545
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_-_15668433 | 0.33 |
ENSDART00000115022
|
zgc:66474
|
zgc:66474 |
| chr22_+_22437561 | 0.32 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr7_-_72423666 | 0.32 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr19_+_46158078 | 0.32 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr14_-_49992709 | 0.32 |
ENSDART00000159988
|
fam193b
|
family with sequence similarity 193, member B |
| chr4_+_90048 | 0.31 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr17_+_43013171 | 0.30 |
ENSDART00000055541
|
gskip
|
gsk3b interacting protein |
| chr3_-_15667713 | 0.30 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr24_-_10394277 | 0.29 |
ENSDART00000127568
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr21_+_30673183 | 0.29 |
ENSDART00000144652
|
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr9_-_29039506 | 0.28 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr8_+_15251448 | 0.28 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr10_-_40352250 | 0.27 |
ENSDART00000150821
|
taar20f
|
trace amine associated receptor 20f |
| chr18_+_1615 | 0.27 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr21_+_6197223 | 0.26 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr10_-_105100 | 0.26 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr2_-_28682476 | 0.26 |
ENSDART00000190178
|
dhcr7
|
7-dehydrocholesterol reductase |
| chr3_-_31845816 | 0.25 |
ENSDART00000157028
|
map3k3
|
mitogen-activated protein kinase kinase kinase 3 |
| chr22_-_881080 | 0.25 |
ENSDART00000185489
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr14_-_30876299 | 0.24 |
ENSDART00000180305
|
ubl3b
|
ubiquitin-like 3b |
| chr22_+_17536989 | 0.24 |
ENSDART00000149531
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr25_-_19661198 | 0.23 |
ENSDART00000149641
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr10_+_589501 | 0.22 |
ENSDART00000188415
|
LO018557.1
|
|
| chr10_+_15106808 | 0.22 |
ENSDART00000141588
|
scpp8
|
secretory calcium-binding phosphoprotein 8 |
| chr5_-_43805597 | 0.21 |
ENSDART00000127956
ENSDART00000028099 |
smn1
|
survival of motor neuron 1, telomeric |
| chr25_-_21085661 | 0.21 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr11_+_19060278 | 0.20 |
ENSDART00000164294
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr22_-_10152629 | 0.20 |
ENSDART00000144209
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr4_+_76466751 | 0.20 |
ENSDART00000164709
|
zgc:153116
|
zgc:153116 |
| chr13_-_24874950 | 0.19 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr10_-_1180314 | 0.19 |
ENSDART00000114261
|
bmpr1bb
|
bone morphogenetic protein receptor, type IBb |
| chr8_-_20838342 | 0.19 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr25_-_13659249 | 0.19 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr17_+_9310259 | 0.19 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr8_-_23765207 | 0.18 |
ENSDART00000141871
|
INAVA
|
si:ch211-163l21.4 |
| chr12_+_30368145 | 0.18 |
ENSDART00000153454
|
ccdc186
|
si:ch211-225b10.4 |
| chr24_+_21514283 | 0.16 |
ENSDART00000007066
|
cdk8
|
cyclin-dependent kinase 8 |
| chr24_-_2917540 | 0.14 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr10_+_44940693 | 0.12 |
ENSDART00000157515
|
cnnm4a
|
cyclin and CBS domain divalent metal cation transport mediator 4a |
| chr25_-_35113891 | 0.11 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr3_-_7129057 | 0.10 |
ENSDART00000125947
|
BX005085.1
|
|
| chr15_-_3078600 | 0.09 |
ENSDART00000172842
|
fndc3a
|
fibronectin type III domain containing 3A |
| chr8_+_23521974 | 0.09 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr19_-_19556778 | 0.09 |
ENSDART00000164060
|
tax1bp1a
|
Tax1 (human T-cell leukemia virus type I) binding protein 1a |
| chr9_-_4598883 | 0.09 |
ENSDART00000171927
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr14_+_6535426 | 0.07 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr16_-_10316359 | 0.07 |
ENSDART00000104025
|
flot1b
|
flotillin 1b |
| chr13_+_45524475 | 0.06 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr7_+_73649686 | 0.06 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr23_+_16430559 | 0.06 |
ENSDART00000112436
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr6_+_8626427 | 0.05 |
ENSDART00000193660
|
tspeara
|
thrombospondin-type laminin G domain and EAR repeats a |
| chr11_-_40101246 | 0.05 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr6_+_3166941 | 0.05 |
ENSDART00000168912
|
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr1_+_55452892 | 0.05 |
ENSDART00000122508
|
CR788255.1
|
|
| chr8_-_37391372 | 0.04 |
ENSDART00000190407
ENSDART00000009569 |
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr17_-_4318393 | 0.04 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr9_+_53719230 | 0.04 |
ENSDART00000165827
|
PCDH8
|
si:ch211-199f5.1 |
| chr24_-_33873451 | 0.04 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr3_-_6768905 | 0.03 |
ENSDART00000193638
ENSDART00000123809 ENSDART00000189440 ENSDART00000188335 |
mast1b
|
microtubule associated serine/threonine kinase 1b |
| chr9_+_41690153 | 0.03 |
ENSDART00000100226
|
ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr8_+_8936912 | 0.03 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr2_-_51454868 | 0.03 |
ENSDART00000172550
|
si:dkeyp-104b3.2
|
si:dkeyp-104b3.2 |
| chr20_+_1834262 | 0.03 |
ENSDART00000185316
|
CABZ01065423.1
|
|
| chr18_+_50278858 | 0.03 |
ENSDART00000014582
|
si:dkey-105e17.1
|
si:dkey-105e17.1 |
| chr21_+_21791799 | 0.03 |
ENSDART00000151759
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr1_+_57176256 | 0.03 |
ENSDART00000152741
|
si:dkey-27j5.6
|
si:dkey-27j5.6 |
| chr3_+_49043917 | 0.03 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr2_+_58221163 | 0.02 |
ENSDART00000157939
|
FO704813.1
|
|
| chr21_+_40261187 | 0.02 |
ENSDART00000142204
|
si:ch211-218m3.11
|
si:ch211-218m3.11 |
| chr13_+_41819817 | 0.02 |
ENSDART00000185778
|
CABZ01066611.1
|
|
| chr5_+_50371951 | 0.01 |
ENSDART00000184488
ENSDART00000171295 |
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr9_-_30160897 | 0.01 |
ENSDART00000143986
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr22_+_15336752 | 0.01 |
ENSDART00000139070
|
sult3st2
|
sulfotransferase family 3, cytosolic sulfotransferase 2 |
| chr1_+_19332837 | 0.01 |
ENSDART00000078594
|
tyrp1b
|
tyrosinase-related protein 1b |
| chr6_-_39051319 | 0.01 |
ENSDART00000155093
|
tns2b
|
tensin 2b |
| chr1_+_56955349 | 0.01 |
ENSDART00000152758
|
si:ch211-1f22.10
|
si:ch211-1f22.10 |
| chr12_+_46740584 | 0.00 |
ENSDART00000171563
|
plaub
|
plasminogen activator, urokinase b |
| chr7_-_17412559 | 0.00 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr15_+_31471808 | 0.00 |
ENSDART00000110078
|
or102-3
|
odorant receptor, family C, subfamily 102, member 3 |
| chr6_+_8129543 | 0.00 |
ENSDART00000011724
|
klf1
|
Kruppel-like factor 1 (erythroid) |
| chr15_-_37921998 | 0.00 |
ENSDART00000193597
ENSDART00000181443 ENSDART00000168790 |
si:dkey-238d18.5
|
si:dkey-238d18.5 |
| chr4_-_61406364 | 0.00 |
ENSDART00000142592
|
znf1021
|
zinc finger protein 1021 |
Network of associatons between targets according to the STRING database.
First level regulatory network of insm1a+insm1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.4 | 1.8 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 1.6 | GO:0040016 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.2 | 0.8 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.1 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 1.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.2 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.4 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.0 | 0.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.2 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 1.0 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.6 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.3 | GO:0030814 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.0 | 0.1 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.9 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.7 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.0 | 0.3 | GO:0042446 | hormone biosynthetic process(GO:0042446) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.5 | GO:0016604 | nuclear body(GO:0016604) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 1.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 1.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.5 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 1.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.7 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.4 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |