Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
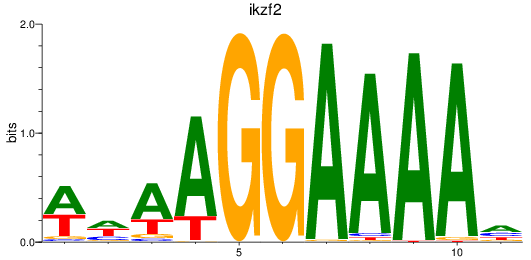
Results for ikzf2
Z-value: 1.11
Transcription factors associated with ikzf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ikzf2
|
ENSDARG00000069111 | IKAROS family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IKZF2 | dr11_v1_chr9_-_40073255_40073255 | 0.41 | 9.4e-02 | Click! |
Activity profile of ikzf2 motif
Sorted Z-values of ikzf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_38626926 | 2.46 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr6_+_38626684 | 2.25 |
ENSDART00000086533
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr19_+_41479990 | 2.15 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr14_-_21618005 | 2.00 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr14_-_34605607 | 1.96 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr17_+_32531854 | 1.95 |
ENSDART00000123399
|
gcfc2
|
GC-rich sequence DNA-binding factor 2 |
| chr16_-_38333976 | 1.81 |
ENSDART00000031895
|
cdc42se1
|
CDC42 small effector 1 |
| chr3_-_15470944 | 1.79 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr14_-_46198373 | 1.78 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr4_+_9279784 | 1.73 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr3_+_30921246 | 1.70 |
ENSDART00000076850
|
cldni
|
claudin i |
| chr14_-_34605804 | 1.69 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr13_-_21672131 | 1.67 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr3_-_25055795 | 1.61 |
ENSDART00000156459
|
ep300b
|
E1A binding protein p300 b |
| chr20_-_44090624 | 1.61 |
ENSDART00000048978
ENSDART00000082283 ENSDART00000082276 |
runx2b
|
runt-related transcription factor 2b |
| chr5_-_28968964 | 1.58 |
ENSDART00000184936
ENSDART00000016628 |
fam129bb
|
family with sequence similarity 129, member Bb |
| chr19_-_18136410 | 1.56 |
ENSDART00000012352
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr20_+_46040666 | 1.53 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr1_-_59169815 | 1.47 |
ENSDART00000100163
|
wu:fk65c09
|
wu:fk65c09 |
| chr15_-_17024779 | 1.45 |
ENSDART00000154719
|
hip1
|
huntingtin interacting protein 1 |
| chr19_+_27859546 | 1.43 |
ENSDART00000161908
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr12_+_33403694 | 1.41 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr17_-_8592824 | 1.40 |
ENSDART00000127022
|
CU462878.1
|
|
| chr16_-_17345377 | 1.39 |
ENSDART00000143056
|
zyx
|
zyxin |
| chr23_+_27782071 | 1.38 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr10_+_33573838 | 1.38 |
ENSDART00000051197
ENSDART00000130093 |
c10h21orf59
|
c10h21orf59 homolog (H. sapiens) |
| chr21_-_19918286 | 1.34 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_-_32876280 | 1.33 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr23_+_40133136 | 1.29 |
ENSDART00000157616
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr25_-_18948816 | 1.28 |
ENSDART00000091549
|
nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr12_+_27232173 | 1.27 |
ENSDART00000193714
|
tmem106a
|
transmembrane protein 106A |
| chr24_-_26622423 | 1.24 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr1_+_14454663 | 1.23 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr8_-_22538588 | 1.23 |
ENSDART00000144041
|
csde1
|
cold shock domain containing E1, RNA-binding |
| chr5_-_50084310 | 1.21 |
ENSDART00000074599
ENSDART00000189970 |
fam172a
|
family with sequence similarity 172, member A |
| chr6_+_28208973 | 1.21 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr8_-_22539113 | 1.21 |
ENSDART00000183297
ENSDART00000185981 |
csde1
|
cold shock domain containing E1, RNA-binding |
| chr12_+_17603528 | 1.20 |
ENSDART00000111565
|
pms2
|
PMS1 homolog 2, mismatch repair system component |
| chr3_+_26245731 | 1.20 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr10_+_36662640 | 1.18 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr8_+_47683539 | 1.17 |
ENSDART00000190701
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr3_-_36127234 | 1.17 |
ENSDART00000130917
|
coil
|
coilin p80 |
| chr12_+_16953415 | 1.16 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr3_+_35498119 | 1.16 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr11_-_21404358 | 1.13 |
ENSDART00000129062
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr5_-_31772559 | 1.12 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr19_-_47832853 | 1.11 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr6_-_10034145 | 1.10 |
ENSDART00000185999
|
nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr22_-_17653143 | 1.10 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr1_+_51391082 | 1.09 |
ENSDART00000063936
|
atg4da
|
autophagy related 4D, cysteine peptidase a |
| chr8_+_21254192 | 1.07 |
ENSDART00000167718
|
itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr6_+_38896158 | 1.05 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr13_+_15580758 | 1.05 |
ENSDART00000087194
ENSDART00000013525 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr18_+_11858397 | 1.03 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr1_+_16548733 | 1.03 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr8_+_47683352 | 1.03 |
ENSDART00000187320
ENSDART00000192605 |
dpp9
|
dipeptidyl-peptidase 9 |
| chr8_-_7232413 | 1.02 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr1_+_35494837 | 1.02 |
ENSDART00000140724
|
gab1
|
GRB2-associated binding protein 1 |
| chr1_+_35495368 | 1.01 |
ENSDART00000053806
|
gab1
|
GRB2-associated binding protein 1 |
| chr19_+_24324967 | 1.01 |
ENSDART00000090081
|
sema4ab
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ab |
| chr20_+_26943072 | 1.01 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr21_+_31253048 | 1.01 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr18_-_12295092 | 1.00 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr23_-_26227805 | 1.00 |
ENSDART00000158082
|
BX927204.1
|
|
| chr19_+_42071814 | 1.00 |
ENSDART00000166422
ENSDART00000191058 |
nfyc
|
nuclear transcription factor Y, gamma |
| chr25_-_12906872 | 1.00 |
ENSDART00000165156
ENSDART00000167449 |
sept15
|
septin 15 |
| chr3_-_34528306 | 0.97 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr6_+_23809163 | 0.96 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr23_+_40139765 | 0.96 |
ENSDART00000185376
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr14_-_24277805 | 0.95 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr20_-_14718801 | 0.95 |
ENSDART00000137605
|
suco
|
SUN domain containing ossification factor |
| chr10_+_7703251 | 0.94 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr3_+_53156813 | 0.93 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr11_-_21404044 | 0.93 |
ENSDART00000080116
|
ikbke
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr4_-_4250317 | 0.92 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr7_-_30553588 | 0.91 |
ENSDART00000139546
|
sltm
|
SAFB-like, transcription modulator |
| chr22_-_155627 | 0.91 |
ENSDART00000110807
|
si:ch1073-335m2.2
|
si:ch1073-335m2.2 |
| chr1_+_54737353 | 0.91 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr5_-_65158203 | 0.91 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr3_+_28576173 | 0.89 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr17_+_19626479 | 0.89 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr25_-_17910714 | 0.89 |
ENSDART00000191586
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr18_-_13056801 | 0.89 |
ENSDART00000088908
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr23_-_270847 | 0.88 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr2_+_22416275 | 0.87 |
ENSDART00000185179
ENSDART00000172715 |
pkn2
|
protein kinase N2 |
| chr11_-_6868287 | 0.87 |
ENSDART00000037824
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_-_39518489 | 0.87 |
ENSDART00000185446
|
atf1
|
activating transcription factor 1 |
| chr15_+_23657051 | 0.86 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr4_-_4261673 | 0.86 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr5_+_65086856 | 0.86 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr9_+_44391254 | 0.86 |
ENSDART00000148826
|
ssfa2
|
sperm specific antigen 2 |
| chr4_-_7876005 | 0.85 |
ENSDART00000109252
|
nudt5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr23_+_1181248 | 0.85 |
ENSDART00000170942
|
utrn
|
utrophin |
| chr17_-_15528597 | 0.83 |
ENSDART00000150232
|
fyna
|
FYN proto-oncogene, Src family tyrosine kinase a |
| chr19_+_46222918 | 0.83 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr11_-_6868474 | 0.81 |
ENSDART00000168372
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_-_36046458 | 0.81 |
ENSDART00000061520
|
brk1
|
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr13_+_34690158 | 0.79 |
ENSDART00000182978
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr22_-_3182965 | 0.78 |
ENSDART00000158009
|
lonp1
|
lon peptidase 1, mitochondrial |
| chr19_+_46222428 | 0.78 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr23_-_18668836 | 0.77 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr5_-_13766651 | 0.77 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr4_+_13909398 | 0.77 |
ENSDART00000187959
ENSDART00000184926 |
pphln1
|
periphilin 1 |
| chr3_-_1263047 | 0.77 |
ENSDART00000184388
|
tcf20
|
transcription factor 20 |
| chr21_+_37513058 | 0.76 |
ENSDART00000141096
|
amot
|
angiomotin |
| chr21_+_25120546 | 0.76 |
ENSDART00000149507
|
ddx10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr25_-_29074064 | 0.74 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr3_+_26244353 | 0.73 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr9_-_28939796 | 0.73 |
ENSDART00000101269
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr2_-_21819421 | 0.73 |
ENSDART00000121586
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_-_26498196 | 0.73 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr15_+_29408886 | 0.72 |
ENSDART00000184581
ENSDART00000184478 |
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr9_-_12659140 | 0.72 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr20_-_18789543 | 0.72 |
ENSDART00000182240
|
ccm2
|
cerebral cavernous malformation 2 |
| chr4_-_18416566 | 0.71 |
ENSDART00000033717
|
cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_-_48603471 | 0.70 |
ENSDART00000189027
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr19_-_20403507 | 0.69 |
ENSDART00000052603
ENSDART00000137590 |
dazl
|
deleted in azoospermia-like |
| chr2_-_52550135 | 0.69 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr6_-_47246948 | 0.69 |
ENSDART00000162435
|
grm4
|
glutamate receptor, metabotropic 4 |
| chr3_-_60856157 | 0.69 |
ENSDART00000053502
|
CABZ01087513.1
|
|
| chr22_+_15720381 | 0.68 |
ENSDART00000128149
|
fam32a
|
family with sequence similarity 32, member A |
| chr15_-_33807758 | 0.68 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr4_-_11053543 | 0.68 |
ENSDART00000067262
|
mettl25
|
methyltransferase like 25 |
| chr20_-_43750771 | 0.67 |
ENSDART00000100605
|
ttc32
|
tetratricopeptide repeat domain 32 |
| chr9_-_55946377 | 0.67 |
ENSDART00000161536
ENSDART00000169432 |
SH3RF3
|
si:ch211-124n19.2 |
| chr21_-_30166097 | 0.67 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr6_+_19950107 | 0.67 |
ENSDART00000181632
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr19_+_32401278 | 0.66 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr16_-_21047483 | 0.65 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr11_+_18216404 | 0.65 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr20_-_40367493 | 0.64 |
ENSDART00000075096
|
smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A |
| chr17_-_11357851 | 0.64 |
ENSDART00000153915
|
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr21_-_36571804 | 0.64 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr3_+_22036113 | 0.64 |
ENSDART00000132190
|
cdc27
|
cell division cycle 27 |
| chr8_-_23599096 | 0.63 |
ENSDART00000183096
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr5_-_23574234 | 0.62 |
ENSDART00000002453
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr19_+_42086862 | 0.62 |
ENSDART00000151605
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr3_+_22035863 | 0.61 |
ENSDART00000177169
|
cdc27
|
cell division cycle 27 |
| chr7_-_64770456 | 0.60 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr5_+_65087226 | 0.60 |
ENSDART00000183187
|
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr19_+_15443540 | 0.60 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr12_-_22379421 | 0.59 |
ENSDART00000187875
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr12_-_17152139 | 0.58 |
ENSDART00000152478
|
stambpl1
|
STAM binding protein-like 1 |
| chr11_+_18873619 | 0.58 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr5_+_63288599 | 0.58 |
ENSDART00000140065
|
si:ch73-37h15.2
|
si:ch73-37h15.2 |
| chr11_+_43740949 | 0.58 |
ENSDART00000189296
|
CU862021.1
|
|
| chr25_+_15273370 | 0.58 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr7_+_31130667 | 0.58 |
ENSDART00000173937
|
tjp1a
|
tight junction protein 1a |
| chr12_+_19036380 | 0.58 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr14_+_20929586 | 0.57 |
ENSDART00000106198
ENSDART00000166366 |
zgc:66433
|
zgc:66433 |
| chr17_-_13058515 | 0.57 |
ENSDART00000172450
ENSDART00000170255 |
CU469462.1
|
|
| chr17_-_8727699 | 0.57 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr24_-_25166720 | 0.57 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr2_-_29994726 | 0.57 |
ENSDART00000163350
|
cnpy1
|
canopy1 |
| chr19_+_46158078 | 0.57 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr23_+_45027263 | 0.56 |
ENSDART00000058364
|
hmgb2b
|
high mobility group box 2b |
| chr10_+_10386435 | 0.56 |
ENSDART00000179214
ENSDART00000189799 ENSDART00000193875 |
sardh
|
sarcosine dehydrogenase |
| chr14_+_45259644 | 0.56 |
ENSDART00000168270
|
CABZ01048053.1
|
|
| chr14_+_46342882 | 0.56 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr16_+_12812214 | 0.56 |
ENSDART00000124875
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr6_+_6662479 | 0.55 |
ENSDART00000065568
|
zgc:113227
|
zgc:113227 |
| chr4_+_279669 | 0.55 |
ENSDART00000184884
|
CABZ01085275.1
|
|
| chr9_-_32684008 | 0.55 |
ENSDART00000041751
|
ercc1
|
excision repair cross-complementation group 1 |
| chr18_-_37252036 | 0.54 |
ENSDART00000132230
|
six5
|
SIX homeobox 5 |
| chr20_+_49787584 | 0.54 |
ENSDART00000193458
ENSDART00000181511 ENSDART00000185850 ENSDART00000185613 ENSDART00000191671 |
CABZ01078261.1
|
|
| chr7_+_24889783 | 0.54 |
ENSDART00000005329
ENSDART00000159955 |
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr12_+_14084291 | 0.54 |
ENSDART00000189734
|
si:ch211-217a12.1
|
si:ch211-217a12.1 |
| chr2_-_5723786 | 0.53 |
ENSDART00000100924
|
cwc25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr24_-_25166416 | 0.53 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr1_+_2712956 | 0.53 |
ENSDART00000126093
|
gpc6a
|
glypican 6a |
| chr13_+_39208542 | 0.53 |
ENSDART00000147971
|
fam135a
|
family with sequence similarity 135, member A |
| chr3_+_36127287 | 0.53 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr1_-_28473350 | 0.52 |
ENSDART00000190608
ENSDART00000148175 |
si:ch1073-440b2.1
|
si:ch1073-440b2.1 |
| chr3_+_12593558 | 0.52 |
ENSDART00000186891
ENSDART00000159252 |
abca3b
|
ATP-binding cassette, sub-family A (ABC1), member 3b |
| chr24_+_14801844 | 0.52 |
ENSDART00000141620
|
pi15a
|
peptidase inhibitor 15a |
| chr11_-_31226578 | 0.52 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr20_+_18943406 | 0.52 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr9_+_12948511 | 0.51 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr12_-_33789006 | 0.51 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr5_-_40024902 | 0.51 |
ENSDART00000017451
|
cds1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr16_+_12812472 | 0.50 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr16_-_42152145 | 0.50 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr9_-_22057658 | 0.50 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr25_+_17860962 | 0.50 |
ENSDART00000163153
|
pth1a
|
parathyroid hormone 1a |
| chr2_-_58183499 | 0.50 |
ENSDART00000172281
ENSDART00000186262 |
si:ch1073-185p12.2
|
si:ch1073-185p12.2 |
| chr14_+_20156477 | 0.49 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr12_-_3778848 | 0.49 |
ENSDART00000152128
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr17_-_5352924 | 0.48 |
ENSDART00000167275
|
supt3h
|
SPT3 homolog, SAGA and STAGA complex component |
| chr5_+_61361815 | 0.48 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr23_+_2717950 | 0.48 |
ENSDART00000137641
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr12_-_33789218 | 0.47 |
ENSDART00000193258
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr16_-_42151909 | 0.47 |
ENSDART00000160950
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr22_+_20169352 | 0.46 |
ENSDART00000169055
ENSDART00000061617 |
hmg20b
|
high mobility group 20B |
| chr11_+_39935154 | 0.46 |
ENSDART00000187653
|
vamp3
|
vesicle-associated membrane protein 3 (cellubrevin) |
| chr8_-_25771474 | 0.46 |
ENSDART00000193883
|
suv39h1b
|
suppressor of variegation 3-9 homolog 1b |
| chr25_-_3808635 | 0.46 |
ENSDART00000075659
ENSDART00000154691 |
gatd1
|
glutamine amidotransferase like class 1 domain containing 1 |
| chr18_+_44703343 | 0.46 |
ENSDART00000131510
|
b3gnt2l
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2, like |
| chr17_-_29213710 | 0.46 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr22_-_14262115 | 0.46 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr24_+_19518570 | 0.45 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ikzf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.4 | 1.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 2.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.2 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 0.9 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 | 1.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.2 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 1.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.2 | 2.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 0.2 | GO:0051295 | establishment of meiotic spindle localization(GO:0051295) |
| 0.2 | 0.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 0.6 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.2 | 1.1 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.2 | 1.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.2 | 1.6 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 0.6 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.2 | 0.9 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 1.4 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.2 | 0.8 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 2.3 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.8 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 4.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.0 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.1 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.7 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.6 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.8 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 1.1 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.8 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.7 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 1.2 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.4 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.8 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 1.7 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.1 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 1.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.8 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.7 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.9 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.4 | GO:0030826 | regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 4.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 1.5 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.1 | 0.4 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.1 | 1.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.5 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.1 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.5 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.6 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.7 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.3 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 1.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.5 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.2 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 2.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 1.4 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 1.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.5 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.7 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 2.4 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.6 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.6 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 2.0 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.0 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.6 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 2.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.7 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.4 | 1.5 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 3.7 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.0 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 2.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.6 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.5 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 1.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 1.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 3.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.9 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.9 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 0.7 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.9 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 1.0 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 4.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.4 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.4 | 1.4 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.6 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 1.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.7 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 0.9 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 1.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 1.5 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.2 | 1.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.2 | 0.6 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 1.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.2 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 1.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.0 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 1.5 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.5 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 1.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 1.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.5 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.9 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 1.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.2 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 1.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.2 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 2.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.5 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.7 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 4.7 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.2 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.9 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0019808 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 3.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.5 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.2 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.4 | GO:0016638 | oxidoreductase activity, acting on the CH-NH2 group of donors(GO:0016638) |
| 0.0 | 0.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.1 | 1.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.7 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.5 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 5.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.9 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 2.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 1.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.9 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |