Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
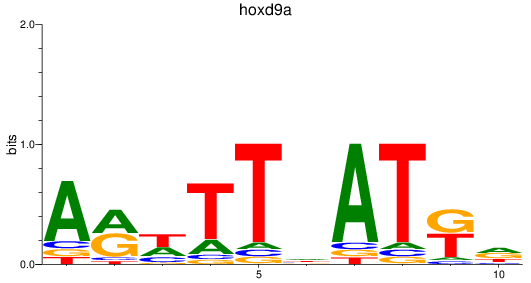
Results for hoxd9a
Z-value: 0.52
Transcription factors associated with hoxd9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd9a
|
ENSDARG00000059274 | homeobox D9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd9a | dr11_v1_chr9_-_1965727_1965727 | 0.56 | 1.5e-02 | Click! |
Activity profile of hoxd9a motif
Sorted Z-values of hoxd9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_20081621 | 0.82 |
ENSDART00000024647
|
dennd6b
|
DENN/MADD domain containing 6B |
| chr3_+_37083765 | 0.79 |
ENSDART00000125611
|
retreg3
|
reticulophagy regulator family member 3 |
| chr7_+_28724919 | 0.78 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr25_+_418932 | 0.75 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr22_-_18241390 | 0.73 |
ENSDART00000083879
|
tmem161a
|
transmembrane protein 161A |
| chr22_+_15959844 | 0.71 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr14_-_16082806 | 0.69 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr21_-_42831033 | 0.69 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr16_-_47427016 | 0.67 |
ENSDART00000074575
|
sept7b
|
septin 7b |
| chr23_+_2666944 | 0.67 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr24_-_3477103 | 0.66 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr12_+_24952902 | 0.63 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr24_-_21090447 | 0.61 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr8_-_39884359 | 0.55 |
ENSDART00000131372
|
mlec
|
malectin |
| chr5_+_43006422 | 0.54 |
ENSDART00000009182
|
aqp3a
|
aquaporin 3a |
| chr13_+_7578111 | 0.53 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr3_+_35005062 | 0.52 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr10_+_16911951 | 0.52 |
ENSDART00000164933
|
UNC13B (1 of many)
|
unc-13 homolog B |
| chr14_+_46342882 | 0.51 |
ENSDART00000193707
ENSDART00000060577 |
tmem33
|
transmembrane protein 33 |
| chr9_+_22780901 | 0.51 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr2_+_30739680 | 0.50 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr1_-_338445 | 0.50 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr11_+_11303458 | 0.48 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr25_+_3099073 | 0.47 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_-_29514689 | 0.45 |
ENSDART00000126018
ENSDART00000125175 |
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr16_+_54209504 | 0.45 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr20_-_51831657 | 0.45 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr16_-_17347727 | 0.45 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr14_-_46617228 | 0.45 |
ENSDART00000161010
ENSDART00000171054 |
prom1a
|
prominin 1a |
| chr22_+_15960005 | 0.45 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr6_-_29051773 | 0.44 |
ENSDART00000190508
ENSDART00000180191 ENSDART00000111682 |
evi5b
|
ecotropic viral integration site 5b |
| chr21_+_3166519 | 0.44 |
ENSDART00000160585
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr25_+_19870603 | 0.43 |
ENSDART00000047251
|
gramd4b
|
GRAM domain containing 4b |
| chr9_-_12269847 | 0.41 |
ENSDART00000136558
ENSDART00000144734 ENSDART00000131766 ENSDART00000032344 |
nup35
|
nucleoporin 35 |
| chr19_-_4137087 | 0.41 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr3_-_46403778 | 0.41 |
ENSDART00000074422
|
cdip1
|
cell death-inducing p53 target 1 |
| chr19_+_1510971 | 0.41 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr15_-_37589600 | 0.41 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr14_+_26224731 | 0.41 |
ENSDART00000168673
|
gm2a
|
GM2 ganglioside activator |
| chr16_-_17586883 | 0.40 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr2_-_57837838 | 0.39 |
ENSDART00000010699
|
sf3a2
|
splicing factor 3a, subunit 2 |
| chr16_+_13883872 | 0.39 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr6_-_2134581 | 0.38 |
ENSDART00000175478
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr24_+_3478871 | 0.38 |
ENSDART00000111491
ENSDART00000134598 ENSDART00000142407 |
wdr37
|
WD repeat domain 37 |
| chr13_-_42757565 | 0.38 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr23_-_14990865 | 0.38 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr21_+_45717930 | 0.38 |
ENSDART00000164315
|
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr4_-_14954029 | 0.38 |
ENSDART00000038642
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr4_-_14954327 | 0.37 |
ENSDART00000182729
|
slc26a5
|
solute carrier family 26 (anion exchanger), member 5 |
| chr23_+_4483083 | 0.37 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr2_-_985417 | 0.37 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr3_+_43774369 | 0.36 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr3_+_39579393 | 0.36 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr9_+_21259820 | 0.36 |
ENSDART00000137024
ENSDART00000132324 |
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr12_-_20616160 | 0.36 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr10_-_10969444 | 0.36 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr3_+_19248973 | 0.36 |
ENSDART00000174668
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr21_+_43172506 | 0.36 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr15_-_1844048 | 0.36 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_-_58901552 | 0.35 |
ENSDART00000122003
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr6_+_21001264 | 0.35 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr16_-_28658341 | 0.35 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr16_-_44945224 | 0.35 |
ENSDART00000156921
|
ncam3
|
neural cell adhesion molecule 3 |
| chr8_+_12951155 | 0.35 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr10_+_44903676 | 0.35 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr11_-_12802123 | 0.34 |
ENSDART00000104143
|
txlng
|
taxilin gamma |
| chr22_-_17671348 | 0.34 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr4_-_1776352 | 0.33 |
ENSDART00000123089
|
dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr24_-_35699595 | 0.33 |
ENSDART00000167990
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_+_28492990 | 0.33 |
ENSDART00000160347
ENSDART00000187176 ENSDART00000088094 |
stag2b
|
stromal antigen 2b |
| chr7_+_9189547 | 0.33 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr13_+_18471546 | 0.33 |
ENSDART00000090712
ENSDART00000127962 |
stox1
|
storkhead box 1 |
| chr4_+_11695979 | 0.32 |
ENSDART00000137736
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr10_+_23060391 | 0.32 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr6_-_8377055 | 0.32 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr18_-_3542435 | 0.31 |
ENSDART00000184017
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr22_+_1947494 | 0.31 |
ENSDART00000159121
|
si:dkey-15h8.15
|
si:dkey-15h8.15 |
| chr20_+_53368611 | 0.31 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr9_+_38883388 | 0.31 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr16_-_10223741 | 0.31 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr24_+_34970680 | 0.31 |
ENSDART00000113014
|
rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr6_-_19333947 | 0.30 |
ENSDART00000160887
|
gga3
|
ggolgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr19_-_31042570 | 0.30 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr15_+_24795473 | 0.30 |
ENSDART00000139689
ENSDART00000141033 ENSDART00000100746 ENSDART00000135677 |
gosr1
|
golgi SNAP receptor complex member 1 |
| chr4_-_12978925 | 0.30 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr11_-_44979281 | 0.30 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr9_-_25366541 | 0.30 |
ENSDART00000021672
|
epc2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr11_-_25461336 | 0.29 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr21_-_22117085 | 0.29 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr17_-_1174960 | 0.29 |
ENSDART00000161202
|
dnajc17
|
DnaJ (Hsp40) homolog, subfamily C, member 17 |
| chr8_+_21133307 | 0.29 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr13_+_15580758 | 0.29 |
ENSDART00000087194
ENSDART00000013525 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr15_-_25435085 | 0.29 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr22_+_34784075 | 0.29 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr13_-_24260609 | 0.29 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr13_+_35689749 | 0.29 |
ENSDART00000158726
|
psme4a
|
proteasome activator subunit 4a |
| chr22_-_11661724 | 0.29 |
ENSDART00000025198
|
mettl21a
|
methyltransferase like 21A |
| chr2_-_10877765 | 0.29 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr16_-_47426482 | 0.28 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr10_-_10969596 | 0.28 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr6_-_8392104 | 0.28 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr2_+_23081402 | 0.28 |
ENSDART00000183073
|
mfsd12a
|
major facilitator superfamily domain containing 12a |
| chr3_+_13848226 | 0.27 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr13_+_35690023 | 0.27 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr16_-_42461263 | 0.27 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr13_+_48359573 | 0.27 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr15_-_26930999 | 0.27 |
ENSDART00000181674
ENSDART00000126046 |
ccdc9
|
coiled-coil domain containing 9 |
| chr19_-_6083761 | 0.26 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr6_-_57539141 | 0.26 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr14_+_23593989 | 0.26 |
ENSDART00000163682
|
med12
|
mediator complex subunit 12 |
| chr24_+_17068724 | 0.26 |
ENSDART00000191137
|
pip4k2aa
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha a |
| chr15_-_14193926 | 0.26 |
ENSDART00000162707
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr2_-_9915814 | 0.26 |
ENSDART00000091644
ENSDART00000177556 |
abi1b
|
abl-interactor 1b |
| chr24_-_15208414 | 0.25 |
ENSDART00000145978
|
rttn
|
rotatin |
| chr12_+_19199735 | 0.25 |
ENSDART00000066393
|
pdap1a
|
pdgfa associated protein 1a |
| chr1_-_69444 | 0.25 |
ENSDART00000166954
|
si:zfos-1011f11.1
|
si:zfos-1011f11.1 |
| chr23_+_36460239 | 0.25 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr11_+_45388126 | 0.24 |
ENSDART00000167826
ENSDART00000171274 |
srsf11
|
serine/arginine-rich splicing factor 11 |
| chr16_-_31505802 | 0.24 |
ENSDART00000147616
|
si:ch211-251p5.5
|
si:ch211-251p5.5 |
| chr24_-_37326236 | 0.24 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr3_-_32873641 | 0.24 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr16_+_26706519 | 0.24 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr19_-_37508571 | 0.24 |
ENSDART00000018255
|
ilf2
|
interleukin enhancer binding factor 2 |
| chr22_+_12595144 | 0.24 |
ENSDART00000140054
ENSDART00000060979 ENSDART00000139826 |
zgc:92335
|
zgc:92335 |
| chr20_+_29565906 | 0.24 |
ENSDART00000062383
|
ywhaqa
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide a |
| chr3_-_31115996 | 0.24 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr5_+_41477526 | 0.24 |
ENSDART00000153567
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr9_-_14683574 | 0.23 |
ENSDART00000144022
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr23_-_7052362 | 0.23 |
ENSDART00000127702
ENSDART00000192468 |
prpf6
|
PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) |
| chr19_+_42061699 | 0.23 |
ENSDART00000125579
|
si:ch211-13c6.2
|
si:ch211-13c6.2 |
| chr3_+_47131112 | 0.23 |
ENSDART00000165100
|
dnm2a
|
dynamin 2a |
| chr21_+_21195487 | 0.22 |
ENSDART00000181746
ENSDART00000184832 |
rictorb
|
RPTOR independent companion of MTOR, complex 2b |
| chr7_-_58098814 | 0.22 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr6_-_40922971 | 0.22 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr23_-_36446307 | 0.22 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr4_+_5317483 | 0.22 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr18_-_16907436 | 0.22 |
ENSDART00000146464
|
swap70b
|
switching B cell complex subunit SWAP70b |
| chr13_-_36535128 | 0.22 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr16_-_25663846 | 0.21 |
ENSDART00000031304
|
derl1
|
derlin 1 |
| chr3_-_31115601 | 0.21 |
ENSDART00000139090
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr6_-_7776612 | 0.21 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr2_-_40889465 | 0.21 |
ENSDART00000192631
ENSDART00000180824 |
uggt1
|
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr3_+_32933663 | 0.20 |
ENSDART00000112742
|
nbr1b
|
neighbor of brca1 gene 1b |
| chr8_+_3820134 | 0.20 |
ENSDART00000122454
|
citb
|
citron rho-interacting serine/threonine kinase b |
| chr25_+_15273370 | 0.20 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr2_-_54387550 | 0.19 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr13_-_24257631 | 0.19 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr20_+_40457599 | 0.18 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr11_-_26701611 | 0.18 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr6_+_11989537 | 0.18 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr9_+_33261330 | 0.18 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr25_-_14433503 | 0.18 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr3_-_9444749 | 0.18 |
ENSDART00000182191
|
FO904885.3
|
|
| chr22_-_22301929 | 0.18 |
ENSDART00000142027
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr8_+_10001805 | 0.17 |
ENSDART00000132894
|
si:dkey-8e10.2
|
si:dkey-8e10.2 |
| chr22_+_9522971 | 0.17 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr19_+_47299212 | 0.17 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr12_+_1286642 | 0.17 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr12_+_9499742 | 0.17 |
ENSDART00000044150
ENSDART00000136354 |
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr23_+_16748806 | 0.16 |
ENSDART00000137737
ENSDART00000142556 |
fbxo44
|
F-box protein 44 |
| chr6_+_19933763 | 0.16 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr22_-_3299100 | 0.16 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr24_-_25166416 | 0.16 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr9_+_17971935 | 0.15 |
ENSDART00000149736
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr2_-_57344037 | 0.15 |
ENSDART00000148873
|
tcf3a
|
transcription factor 3a |
| chr5_-_52964789 | 0.15 |
ENSDART00000166267
|
zfpl1
|
zinc finger protein-like 1 |
| chr25_+_8407892 | 0.15 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr20_-_21910043 | 0.15 |
ENSDART00000152290
ENSDART00000109084 |
daam1b
|
dishevelled associated activator of morphogenesis 1b |
| chr6_-_31348999 | 0.14 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_+_26686279 | 0.14 |
ENSDART00000193543
|
tango2
|
transport and golgi organization 2 homolog (Drosophila) |
| chr4_-_16641742 | 0.14 |
ENSDART00000180605
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr5_-_52010122 | 0.14 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr14_+_41406321 | 0.14 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr3_+_22442445 | 0.14 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr22_-_3299355 | 0.14 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr2_+_10878406 | 0.14 |
ENSDART00000091497
|
tceanc2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2 |
| chr17_+_43595692 | 0.13 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr4_+_75200467 | 0.13 |
ENSDART00000122593
|
CABZ01043955.1
|
|
| chr17_-_13026634 | 0.13 |
ENSDART00000113713
|
fam177a1
|
family with sequence similarity 177, member A1 |
| chr6_-_51771634 | 0.13 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr16_-_10516970 | 0.13 |
ENSDART00000110606
|
abhd16a
|
abhydrolase domain containing 16A |
| chr7_-_7797654 | 0.13 |
ENSDART00000084503
ENSDART00000192779 ENSDART00000173079 |
trmt10b
|
tRNA methyltransferase 10B |
| chr22_-_7778265 | 0.12 |
ENSDART00000097276
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr4_-_15103646 | 0.12 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr24_+_37533728 | 0.12 |
ENSDART00000061203
|
rhot2
|
ras homolog family member T2 |
| chr8_-_36287046 | 0.12 |
ENSDART00000162877
|
si:busm1-194e12.11
|
si:busm1-194e12.11 |
| chr18_-_39473055 | 0.12 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr2_+_45344070 | 0.11 |
ENSDART00000147245
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr6_+_21684296 | 0.11 |
ENSDART00000057223
|
rhebl1
|
Ras homolog, mTORC1 binding like 1 |
| chr4_+_25912308 | 0.11 |
ENSDART00000167845
ENSDART00000136927 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr1_+_46579885 | 0.11 |
ENSDART00000144821
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr18_+_44849809 | 0.11 |
ENSDART00000097328
|
arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr8_+_8238727 | 0.11 |
ENSDART00000144838
|
plxnb3
|
plexin B3 |
| chr5_-_41984298 | 0.11 |
ENSDART00000189535
|
ncor1
|
nuclear receptor corepressor 1 |
| chr16_-_25400257 | 0.11 |
ENSDART00000040756
|
zgc:136493
|
zgc:136493 |
| chr13_-_41155472 | 0.10 |
ENSDART00000160588
|
si:dkeyp-86d6.2
|
si:dkeyp-86d6.2 |
| chr4_+_25912654 | 0.10 |
ENSDART00000109508
ENSDART00000134218 |
vezt
|
vezatin, adherens junctions transmembrane protein |
| chr17_-_45383699 | 0.10 |
ENSDART00000141182
|
tmem206
|
transmembrane protein 206 |
| chr7_+_25135739 | 0.10 |
ENSDART00000167326
|
exoc6b
|
exocyst complex component 6B |
| chr6_-_52675630 | 0.09 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.7 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.4 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 1.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.4 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.6 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.3 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.3 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 0.3 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 0.5 | GO:1901339 | regulation of store-operated calcium channel activity(GO:1901339) |
| 0.1 | 0.3 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.1 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.3 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 0.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.7 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.4 | GO:0006999 | NLS-bearing protein import into nucleus(GO:0006607) nuclear pore organization(GO:0006999) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0052651 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.1 | GO:0036445 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.4 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.4 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 1.0 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.4 | GO:0009636 | response to toxic substance(GO:0009636) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.0 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.9 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 0.4 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.3 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.6 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.8 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.4 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.8 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.6 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |