Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
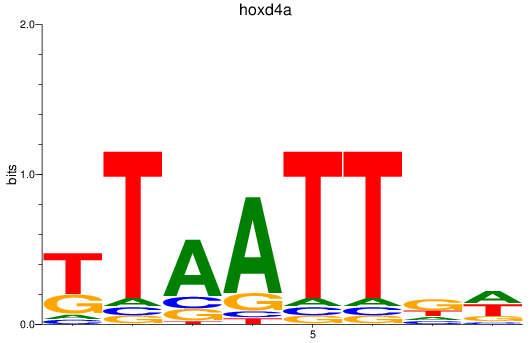
Results for hoxd4a
Z-value: 0.39
Transcription factors associated with hoxd4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd4a
|
ENSDARG00000059276 | homeobox D4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd4a | dr11_v1_chr9_-_1951144_1951144 | 0.37 | 1.3e-01 | Click! |
Activity profile of hoxd4a motif
Sorted Z-values of hoxd4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_31868268 | 0.99 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr25_-_10503043 | 0.86 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr22_+_38037530 | 0.82 |
ENSDART00000012212
|
commd2
|
COMM domain containing 2 |
| chr3_+_24361096 | 0.71 |
ENSDART00000132387
|
pvalb6
|
parvalbumin 6 |
| chr20_+_36628059 | 0.69 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr3_-_49382896 | 0.68 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr9_+_48219111 | 0.67 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr12_-_990149 | 0.63 |
ENSDART00000054367
|
kdelr2b
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2b |
| chr11_+_25259058 | 0.63 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr16_+_26017360 | 0.60 |
ENSDART00000149466
|
prss59.2
|
protease, serine, 59, tandem duplicate 2 |
| chr5_-_61970674 | 0.59 |
ENSDART00000143161
|
znf541
|
zinc finger protein 541 |
| chr2_+_16160906 | 0.59 |
ENSDART00000135783
|
selenoj
|
selenoprotein J |
| chr15_-_45110011 | 0.58 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr12_+_3871452 | 0.55 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr5_+_42400777 | 0.54 |
ENSDART00000183114
|
BX548073.8
|
|
| chr19_-_1855368 | 0.53 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr17_-_39772999 | 0.53 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr6_-_2627488 | 0.53 |
ENSDART00000044089
ENSDART00000158333 ENSDART00000155109 |
hyi
|
hydroxypyruvate isomerase |
| chr2_+_2181709 | 0.52 |
ENSDART00000092763
ENSDART00000148553 |
ccdc13
|
coiled-coil domain containing 13 |
| chr23_+_39695827 | 0.52 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr23_+_24085531 | 0.49 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr1_+_50921266 | 0.49 |
ENSDART00000006538
|
otx2a
|
orthodenticle homeobox 2a |
| chr23_-_42752387 | 0.46 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr5_-_18924834 | 0.45 |
ENSDART00000041371
|
poli
|
polymerase (DNA directed) iota |
| chr1_+_44003654 | 0.45 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr3_+_55031685 | 0.44 |
ENSDART00000132587
|
mpg
|
N-methylpurine DNA glycosylase |
| chr10_-_43771447 | 0.44 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr23_+_19813677 | 0.42 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr5_+_26847190 | 0.42 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr19_+_45962016 | 0.42 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr5_-_19394440 | 0.41 |
ENSDART00000163771
|
foxn4
|
forkhead box N4 |
| chr7_+_65309738 | 0.41 |
ENSDART00000169228
|
vat1l
|
vesicle amine transport 1-like |
| chr20_-_29474859 | 0.41 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr20_-_16171297 | 0.39 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr24_+_37902687 | 0.39 |
ENSDART00000042273
|
ccdc78
|
coiled-coil domain containing 78 |
| chr5_+_23597144 | 0.38 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr20_-_438646 | 0.38 |
ENSDART00000009196
|
ufl1
|
UFM1-specific ligase 1 |
| chr4_+_16885854 | 0.37 |
ENSDART00000017726
|
etnk1
|
ethanolamine kinase 1 |
| chr7_-_59311165 | 0.37 |
ENSDART00000171105
|
m1ap
|
meiosis 1 associated protein |
| chr4_-_74892355 | 0.37 |
ENSDART00000188725
|
PHF21B
|
PHD finger protein 21B |
| chr6_+_49053319 | 0.37 |
ENSDART00000124524
|
sycp1
|
synaptonemal complex protein 1 |
| chr24_+_20536056 | 0.37 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr24_-_37640705 | 0.36 |
ENSDART00000066583
|
zgc:112496
|
zgc:112496 |
| chr2_+_54389750 | 0.36 |
ENSDART00000189236
|
rab12
|
RAB12, member RAS oncogene family |
| chr24_+_21621654 | 0.36 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr19_+_43297546 | 0.35 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr7_+_22657566 | 0.35 |
ENSDART00000141048
|
ponzr5
|
plac8 onzin related protein 5 |
| chr22_+_7439186 | 0.35 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr7_-_41512999 | 0.34 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr15_+_42285643 | 0.33 |
ENSDART00000152731
|
scaf4b
|
SR-related CTD-associated factor 4b |
| chr20_+_30939178 | 0.33 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr4_+_14900042 | 0.32 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr14_-_32089117 | 0.32 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr2_+_33726862 | 0.32 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr17_-_24680965 | 0.32 |
ENSDART00000154880
|
arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr5_+_24882633 | 0.31 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr7_-_27038488 | 0.30 |
ENSDART00000052731
ENSDART00000191382 |
nucb2a
|
nucleobindin 2a |
| chr12_+_20352400 | 0.29 |
ENSDART00000066383
|
hbae5
|
hemoglobin, alpha embryonic 5 |
| chr1_-_45662774 | 0.29 |
ENSDART00000042158
|
serhl
|
serine hydrolase-like |
| chr25_+_18563476 | 0.29 |
ENSDART00000170841
|
cav1
|
caveolin 1 |
| chr7_-_20758825 | 0.28 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr15_-_14884332 | 0.28 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr19_-_47527093 | 0.28 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr18_+_30508729 | 0.28 |
ENSDART00000185140
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr6_+_30689239 | 0.27 |
ENSDART00000065206
|
wdr78
|
WD repeat domain 78 |
| chr9_+_29616854 | 0.26 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr12_+_18899396 | 0.26 |
ENSDART00000105858
|
xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr16_-_41465542 | 0.25 |
ENSDART00000169116
ENSDART00000187446 |
cpne4a
|
copine IVa |
| chr7_+_12927944 | 0.25 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr14_-_7137808 | 0.25 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr15_-_34658057 | 0.25 |
ENSDART00000110964
|
bag6
|
BCL2 associated athanogene 6 |
| chr19_+_22074468 | 0.25 |
ENSDART00000136294
ENSDART00000090476 |
atp9b
|
ATPase phospholipid transporting 9B |
| chr16_+_7242610 | 0.24 |
ENSDART00000081477
|
sri
|
sorcin |
| chr22_+_37631234 | 0.24 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr10_+_39893439 | 0.24 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr2_+_20802876 | 0.24 |
ENSDART00000142321
|
prg4a
|
proteoglycan 4a |
| chr4_-_14915268 | 0.23 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr11_-_41220794 | 0.23 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr18_-_49318823 | 0.23 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr7_-_73851280 | 0.23 |
ENSDART00000190053
|
FP236812.3
|
|
| chr18_+_16192083 | 0.23 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr1_-_46875493 | 0.23 |
ENSDART00000115081
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr24_-_21819010 | 0.23 |
ENSDART00000091096
|
CR352265.1
|
|
| chr15_-_21877726 | 0.22 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr21_-_19314618 | 0.22 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr11_+_1608348 | 0.21 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr15_-_1822548 | 0.21 |
ENSDART00000082026
ENSDART00000180230 |
mmp28
|
matrix metallopeptidase 28 |
| chr14_-_5678457 | 0.20 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr5_-_10239079 | 0.20 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr11_-_22371105 | 0.20 |
ENSDART00000146873
|
tmem183a
|
transmembrane protein 183A |
| chr12_+_28799988 | 0.19 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr12_+_47698356 | 0.19 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr25_+_14165447 | 0.19 |
ENSDART00000145387
|
shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr22_-_19552796 | 0.19 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr22_+_37631034 | 0.19 |
ENSDART00000159016
ENSDART00000193346 |
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr6_+_18569453 | 0.18 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr23_-_44207349 | 0.18 |
ENSDART00000186276
|
zgc:158659
|
zgc:158659 |
| chr8_+_20951590 | 0.18 |
ENSDART00000138728
|
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr2_-_1364678 | 0.17 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr5_+_66433287 | 0.17 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr20_+_10544100 | 0.17 |
ENSDART00000113927
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr8_+_17069577 | 0.17 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr1_-_8192418 | 0.17 |
ENSDART00000136654
|
grapb
|
GRB2-related adaptor protein b |
| chr13_+_45709380 | 0.17 |
ENSDART00000192862
|
si:ch211-62a1.3
|
si:ch211-62a1.3 |
| chr17_+_13088800 | 0.16 |
ENSDART00000149779
|
gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr23_-_16485190 | 0.16 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr1_+_8442071 | 0.16 |
ENSDART00000143547
|
myo15ab
|
myosin XVAb |
| chr23_+_4689626 | 0.16 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr22_+_16555939 | 0.15 |
ENSDART00000012604
|
pdzk1ip1
|
PDZK1 interacting protein 1 |
| chr16_+_3004422 | 0.15 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr3_+_25825043 | 0.15 |
ENSDART00000153749
|
pik3r6b
|
phosphoinositide-3-kinase, regulatory subunit 6b |
| chr23_+_2760573 | 0.15 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr5_+_67268110 | 0.15 |
ENSDART00000148116
|
si:ch211-110p13.9
|
si:ch211-110p13.9 |
| chr25_+_18965430 | 0.15 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr4_-_14315855 | 0.15 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr24_+_26134209 | 0.15 |
ENSDART00000038824
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr9_-_52490579 | 0.15 |
ENSDART00000161667
|
smarcal1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr24_+_38301080 | 0.14 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr20_+_18551657 | 0.14 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr21_-_22689805 | 0.14 |
ENSDART00000157560
ENSDART00000110792 |
gig2e
|
grass carp reovirus (GCRV)-induced gene 2e |
| chr8_-_44004135 | 0.14 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr25_-_24248000 | 0.14 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr11_+_36665359 | 0.14 |
ENSDART00000166144
|
si:ch211-11c3.9
|
si:ch211-11c3.9 |
| chr6_-_43882696 | 0.13 |
ENSDART00000064938
|
foxp1b
|
forkhead box P1b |
| chr4_-_56898328 | 0.13 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr15_-_28082310 | 0.13 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr4_+_16787488 | 0.13 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr23_+_40460333 | 0.13 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr15_-_30816370 | 0.13 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr21_+_9689103 | 0.12 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr15_-_20400423 | 0.12 |
ENSDART00000081290
ENSDART00000156813 ENSDART00000192427 |
rnaset2l
|
ribonuclease T2, like |
| chr2_+_31614533 | 0.12 |
ENSDART00000111038
ENSDART00000181311 |
si:ch211-106h4.6
|
si:ch211-106h4.6 |
| chr7_-_38698583 | 0.12 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr7_+_19495379 | 0.12 |
ENSDART00000180514
|
si:ch211-212k18.8
|
si:ch211-212k18.8 |
| chr16_-_15988320 | 0.12 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr1_+_23784905 | 0.12 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr10_-_44355534 | 0.12 |
ENSDART00000160231
|
sbno1
|
strawberry notch homolog 1 (Drosophila) |
| chr18_-_33979693 | 0.11 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr16_-_13004166 | 0.11 |
ENSDART00000133735
|
cacng7b
|
calcium channel, voltage-dependent, gamma subunit 7b |
| chr10_+_40598791 | 0.11 |
ENSDART00000131895
|
taar17a
|
trace amine associated receptor 17a |
| chr6_-_27891961 | 0.11 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr17_+_32343121 | 0.11 |
ENSDART00000156051
|
dhx32b
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32b |
| chr17_+_25444323 | 0.11 |
ENSDART00000030691
|
clic4
|
chloride intracellular channel 4 |
| chr3_-_39198113 | 0.11 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr5_+_36611128 | 0.11 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr13_-_7567707 | 0.11 |
ENSDART00000190296
ENSDART00000180348 |
pitx3
|
paired-like homeodomain 3 |
| chr15_+_42397125 | 0.10 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr23_-_35491587 | 0.10 |
ENSDART00000153794
|
fam110c
|
family with sequence similarity 110, member C |
| chr14_-_413273 | 0.10 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr13_-_49585385 | 0.10 |
ENSDART00000165868
|
tomm20a
|
translocase of outer mitochondrial membrane 20 |
| chr25_-_18953322 | 0.10 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr22_+_18166660 | 0.10 |
ENSDART00000105432
|
borcs8
|
BLOC-1 related complex subunit 8 |
| chr25_-_13839743 | 0.09 |
ENSDART00000158780
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr4_-_43388943 | 0.09 |
ENSDART00000150796
|
si:dkey-29j8.2
|
si:dkey-29j8.2 |
| chr15_+_23799461 | 0.09 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr10_+_40578332 | 0.09 |
ENSDART00000146155
|
si:ch211-238p8.18
|
si:ch211-238p8.18 |
| chr6_-_39270851 | 0.09 |
ENSDART00000148839
|
arhgef25b
|
Rho guanine nucleotide exchange factor (GEF) 25b |
| chr20_-_29633507 | 0.09 |
ENSDART00000040292
|
cpsf3
|
cleavage and polyadenylation specific factor 3 |
| chr5_-_30491172 | 0.09 |
ENSDART00000138464
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr5_-_66768121 | 0.09 |
ENSDART00000141095
|
FERMT3 (1 of many)
|
im:7154036 |
| chr16_-_55028740 | 0.08 |
ENSDART00000156368
ENSDART00000161704 |
zgc:114181
|
zgc:114181 |
| chr7_+_48761875 | 0.08 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr6_+_41191482 | 0.08 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr1_-_999556 | 0.08 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr23_-_21453614 | 0.08 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr11_-_41996957 | 0.08 |
ENSDART00000055706
|
her15.2
|
hairy and enhancer of split-related 15, tandem duplicate 2 |
| chr11_-_27874116 | 0.08 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr2_+_20793982 | 0.08 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr7_+_20017211 | 0.08 |
ENSDART00000100808
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr23_+_23182037 | 0.08 |
ENSDART00000137353
|
klhl17
|
kelch-like family member 17 |
| chr7_-_17570923 | 0.07 |
ENSDART00000188476
ENSDART00000080624 |
nitr5
|
novel immune-type receptor 5 |
| chr25_+_3318192 | 0.07 |
ENSDART00000146154
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr18_-_30020879 | 0.07 |
ENSDART00000162086
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr19_+_7757682 | 0.07 |
ENSDART00000092138
ENSDART00000092112 |
ubap2l
|
ubiquitin associated protein 2-like |
| chr7_-_9873652 | 0.07 |
ENSDART00000006343
|
asb7
|
ankyrin repeat and SOCS box containing 7 |
| chr7_-_24699985 | 0.07 |
ENSDART00000052802
|
calb2b
|
calbindin 2b |
| chr1_+_50987535 | 0.07 |
ENSDART00000140657
|
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr21_-_13123176 | 0.07 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr15_-_17813680 | 0.07 |
ENSDART00000158556
|
CT573342.2
|
|
| chr10_+_39952995 | 0.07 |
ENSDART00000183077
|
BX927333.1
|
|
| chr10_-_41980797 | 0.07 |
ENSDART00000076575
|
rhof
|
ras homolog family member F |
| chr16_-_17578503 | 0.07 |
ENSDART00000122212
|
casp2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr14_+_4807207 | 0.07 |
ENSDART00000167145
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr11_+_10975012 | 0.06 |
ENSDART00000192872
|
itgb6
|
integrin, beta 6 |
| chr2_+_37815687 | 0.06 |
ENSDART00000166352
|
sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr23_+_404975 | 0.06 |
ENSDART00000181336
|
CU929146.1
|
|
| chr16_-_17541890 | 0.06 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr5_+_59392183 | 0.06 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr13_+_30692669 | 0.06 |
ENSDART00000187818
|
CR762483.1
|
|
| chr4_-_815871 | 0.06 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr20_+_15600167 | 0.06 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr25_+_30074947 | 0.06 |
ENSDART00000154849
|
si:ch211-147k10.5
|
si:ch211-147k10.5 |
| chr12_+_18681477 | 0.06 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr4_+_28364237 | 0.06 |
ENSDART00000187933
ENSDART00000150424 |
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr7_+_5910467 | 0.06 |
ENSDART00000173232
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr6_-_8480815 | 0.06 |
ENSDART00000162300
|
rasal3
|
RAS protein activator like 3 |
| chr4_-_18202291 | 0.06 |
ENSDART00000169704
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr22_-_24757785 | 0.06 |
ENSDART00000078225
|
vtg5
|
vitellogenin 5 |
| chr23_-_29553430 | 0.05 |
ENSDART00000157773
ENSDART00000126384 |
ube4b
|
ubiquitination factor E4B, UFD2 homolog (S. cerevisiae) |
| chr8_+_28065803 | 0.05 |
ENSDART00000178481
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.4 | GO:1990592 | IRE1-mediated unfolded protein response(GO:0036498) negative regulation of response to endoplasmic reticulum stress(GO:1903573) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.5 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.2 | GO:0042822 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.3 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 0.3 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.2 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.3 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.3 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.7 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0071435 | potassium ion export(GO:0071435) potassium ion export across plasma membrane(GO:0097623) |
| 0.0 | 0.1 | GO:0046083 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.7 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.3 | GO:0042744 | oxygen transport(GO:0015671) hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.1 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.2 | 0.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.4 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.3 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 0.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 1.1 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.6 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 1.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.4 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |