Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
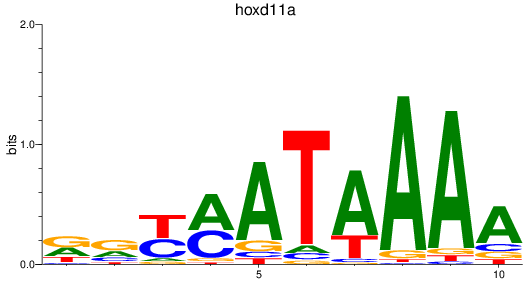
Results for hoxd11a
Z-value: 0.54
Transcription factors associated with hoxd11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd11a
|
ENSDARG00000059267 | homeobox D11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxd11a | dr11_v1_chr9_-_1978090_1978090 | -0.38 | 1.2e-01 | Click! |
Activity profile of hoxd11a motif
Sorted Z-values of hoxd11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_28208973 | 1.61 |
ENSDART00000171216
ENSDART00000171377 ENSDART00000167389 ENSDART00000166988 |
LSM2 (1 of many)
|
si:ch73-14h10.2 |
| chr2_-_32738535 | 1.55 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr23_+_2666944 | 1.50 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr5_+_44805028 | 1.46 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr9_+_33154841 | 1.37 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr5_+_44805269 | 1.28 |
ENSDART00000136965
|
ctsla
|
cathepsin La |
| chr2_+_16597011 | 1.22 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr14_+_32918172 | 1.21 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr11_-_25257045 | 1.17 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr20_-_49889111 | 1.04 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr15_-_44052927 | 0.99 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr17_+_14965570 | 0.99 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr3_-_26183699 | 0.99 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr7_-_24112484 | 0.97 |
ENSDART00000111923
|
ajuba
|
ajuba LIM protein |
| chr19_-_47571456 | 0.95 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr20_-_34670236 | 0.95 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_-_3366590 | 0.94 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr20_-_29499363 | 0.93 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr21_-_19918286 | 0.93 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_+_34490445 | 0.91 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr24_+_21720304 | 0.91 |
ENSDART00000147250
ENSDART00000048273 |
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr5_-_65662996 | 0.90 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr15_-_17024779 | 0.89 |
ENSDART00000154719
|
hip1
|
huntingtin interacting protein 1 |
| chr20_-_51831657 | 0.87 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr20_-_48701593 | 0.86 |
ENSDART00000132835
|
pax1b
|
paired box 1b |
| chr20_-_34028967 | 0.85 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr8_-_25033681 | 0.84 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr20_-_51831816 | 0.82 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr24_+_29912509 | 0.81 |
ENSDART00000168422
|
frrs1b
|
ferric-chelate reductase 1b |
| chr11_-_25257595 | 0.80 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr3_-_26184018 | 0.78 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr2_+_49805892 | 0.78 |
ENSDART00000056248
|
wdr48b
|
WD repeat domain 48b |
| chr16_+_38940758 | 0.76 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr1_+_44173245 | 0.74 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr19_-_47571797 | 0.73 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_+_39579393 | 0.70 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_+_13031497 | 0.70 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr15_+_29024895 | 0.69 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr10_-_10969596 | 0.67 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr5_-_29531948 | 0.66 |
ENSDART00000098360
|
arrdc1a
|
arrestin domain containing 1a |
| chr3_-_30488063 | 0.66 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr17_+_17764979 | 0.65 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr6_-_37745508 | 0.64 |
ENSDART00000078316
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr8_-_2616326 | 0.64 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr20_-_14462995 | 0.63 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr11_+_19603251 | 0.63 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr7_+_38809241 | 0.62 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr1_+_52481332 | 0.62 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr18_-_12295092 | 0.61 |
ENSDART00000033248
|
fam107b
|
family with sequence similarity 107, member B |
| chr16_-_48428035 | 0.61 |
ENSDART00000190554
|
rad21a
|
RAD21 cohesin complex component a |
| chr2_-_44777592 | 0.61 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr7_+_10911396 | 0.60 |
ENSDART00000167273
ENSDART00000081323 ENSDART00000170655 |
abhd17c
|
abhydrolase domain containing 17C |
| chr14_+_45028062 | 0.60 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr5_-_36597612 | 0.57 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr6_-_33685325 | 0.57 |
ENSDART00000181883
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr9_+_21977383 | 0.57 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr6_-_42388608 | 0.54 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr15_-_16946124 | 0.54 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr23_+_22785375 | 0.54 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr10_-_10969444 | 0.53 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr21_-_39024754 | 0.53 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr16_-_42965192 | 0.52 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr9_+_38588081 | 0.52 |
ENSDART00000031127
ENSDART00000131784 |
snx4
|
sorting nexin 4 |
| chr11_+_24313931 | 0.52 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr22_+_10713713 | 0.51 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr16_-_28658341 | 0.50 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr24_-_32025637 | 0.49 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr13_-_37474989 | 0.49 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr18_+_15876385 | 0.49 |
ENSDART00000142527
|
eea1
|
early endosome antigen 1 |
| chr22_+_8979955 | 0.48 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr10_+_5234327 | 0.47 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr2_+_31948352 | 0.46 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr12_-_13650344 | 0.45 |
ENSDART00000124364
ENSDART00000124638 ENSDART00000171929 |
stat5b
|
signal transducer and activator of transcription 5b |
| chr8_+_13364950 | 0.44 |
ENSDART00000159760
|
slc5a5
|
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
| chr14_+_4276394 | 0.44 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr17_-_29311835 | 0.43 |
ENSDART00000104224
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr2_+_22363824 | 0.41 |
ENSDART00000163172
|
hs2st1a
|
heparan sulfate 2-O-sulfotransferase 1a |
| chr18_-_39188664 | 0.41 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr13_-_25745089 | 0.40 |
ENSDART00000189333
ENSDART00000147420 |
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr5_+_20453874 | 0.40 |
ENSDART00000124545
ENSDART00000008402 |
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr24_-_36238054 | 0.40 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr3_+_52545400 | 0.38 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr8_+_36582728 | 0.38 |
ENSDART00000049230
|
pqbp1
|
polyglutamine binding protein 1 |
| chr21_-_11970199 | 0.37 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr5_+_50913034 | 0.37 |
ENSDART00000149787
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr16_-_21903083 | 0.37 |
ENSDART00000165849
|
setdb1b
|
SET domain, bifurcated 1b |
| chr17_+_23554932 | 0.37 |
ENSDART00000135814
|
pank1a
|
pantothenate kinase 1a |
| chr8_-_35960987 | 0.37 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr15_+_44093286 | 0.36 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr10_-_25328814 | 0.36 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr3_+_52545014 | 0.35 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr25_-_34973211 | 0.34 |
ENSDART00000045177
|
cdk10
|
cyclin-dependent kinase 10 |
| chr7_-_26076970 | 0.34 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr20_+_25879826 | 0.33 |
ENSDART00000018519
|
zgc:153896
|
zgc:153896 |
| chr19_+_9455218 | 0.33 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr3_-_31115601 | 0.33 |
ENSDART00000139090
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr21_-_33032715 | 0.33 |
ENSDART00000065349
ENSDART00000191502 ENSDART00000149386 |
slc34a1b
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1b |
| chr25_-_6389713 | 0.33 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr8_+_44677569 | 0.33 |
ENSDART00000191104
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr23_+_43718115 | 0.33 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr5_-_62940851 | 0.33 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr10_+_26990095 | 0.32 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr24_+_11908480 | 0.31 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr10_+_20392656 | 0.31 |
ENSDART00000160803
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr10_-_36618674 | 0.31 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr5_-_57289872 | 0.30 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr23_-_31969786 | 0.30 |
ENSDART00000134550
|
ormdl2
|
ORMDL sphingolipid biosynthesis regulator 2 |
| chr13_-_37465524 | 0.29 |
ENSDART00000100324
ENSDART00000147336 |
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr24_+_11908833 | 0.29 |
ENSDART00000178622
|
fen1
|
flap structure-specific endonuclease 1 |
| chr22_+_1953096 | 0.29 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr5_-_23715861 | 0.29 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr3_-_31115996 | 0.29 |
ENSDART00000058955
|
arl6ip1
|
ADP-ribosylation factor-like 6 interacting protein 1 |
| chr21_-_32781612 | 0.28 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr25_+_33063762 | 0.28 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr12_+_23912074 | 0.28 |
ENSDART00000152864
|
svila
|
supervillin a |
| chr23_-_43718067 | 0.27 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr6_-_31348999 | 0.27 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr20_-_25709247 | 0.27 |
ENSDART00000146711
|
si:dkeyp-117h8.2
|
si:dkeyp-117h8.2 |
| chr7_-_7692992 | 0.26 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr22_-_10158038 | 0.26 |
ENSDART00000047444
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_-_37881345 | 0.26 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr17_+_25519089 | 0.25 |
ENSDART00000041721
|
aim1a
|
crystallin beta-gamma domain containing 1a |
| chr7_-_7692723 | 0.25 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr1_+_2321384 | 0.25 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr1_-_20593778 | 0.25 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr14_+_30795559 | 0.23 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr2_+_23039041 | 0.23 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr7_+_13639473 | 0.23 |
ENSDART00000157870
ENSDART00000032899 |
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr8_-_10961991 | 0.23 |
ENSDART00000139603
|
trim33
|
tripartite motif containing 33 |
| chr8_-_38317914 | 0.23 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_26105141 | 0.22 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr15_-_25527580 | 0.22 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr17_+_15788100 | 0.21 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr20_+_41906960 | 0.20 |
ENSDART00000193460
|
cep85l
|
centrosomal protein 85, like |
| chr17_-_15498275 | 0.19 |
ENSDART00000156905
ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr24_+_40529346 | 0.19 |
ENSDART00000168548
|
CU929259.1
|
|
| chr16_+_54829293 | 0.19 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr8_+_40477264 | 0.18 |
ENSDART00000085559
|
gck
|
glucokinase (hexokinase 4) |
| chr12_-_20658285 | 0.17 |
ENSDART00000080888
|
snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr8_+_7740004 | 0.17 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr25_+_28823952 | 0.17 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr2_-_32366287 | 0.17 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr4_-_16836006 | 0.16 |
ENSDART00000010777
|
ldhba
|
lactate dehydrogenase Ba |
| chr8_+_3379815 | 0.15 |
ENSDART00000155995
|
FUT9 (1 of many)
|
zgc:136963 |
| chr15_+_29727799 | 0.15 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr18_-_15373620 | 0.15 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr11_-_36009924 | 0.15 |
ENSDART00000189959
ENSDART00000167472 ENSDART00000191211 ENSDART00000191662 ENSDART00000191780 ENSDART00000192622 ENSDART00000179911 |
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr22_+_2632961 | 0.15 |
ENSDART00000106278
|
znf1173
|
zinc finger protein 1173 |
| chr14_+_3071110 | 0.14 |
ENSDART00000162445
|
hmgxb3
|
HMG box domain containing 3 |
| chr17_-_44249720 | 0.14 |
ENSDART00000156648
|
otx2b
|
orthodenticle homeobox 2b |
| chr2_+_35733335 | 0.14 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr20_-_39367895 | 0.13 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr8_+_23521974 | 0.13 |
ENSDART00000188130
ENSDART00000129378 |
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr2_-_40135942 | 0.12 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr4_-_22338906 | 0.12 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr14_+_36497250 | 0.12 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr16_+_54829574 | 0.11 |
ENSDART00000148392
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr25_+_18953575 | 0.11 |
ENSDART00000155610
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr6_+_13083146 | 0.11 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr5_-_28767573 | 0.10 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr25_+_18953756 | 0.10 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr18_+_14342326 | 0.10 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr12_+_34854562 | 0.09 |
ENSDART00000130366
|
si:dkey-21c1.4
|
si:dkey-21c1.4 |
| chr11_+_37049347 | 0.09 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr17_+_8799661 | 0.09 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_-_15559817 | 0.09 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr19_-_14191592 | 0.08 |
ENSDART00000164594
|
tbxta
|
T-box transcription factor Ta |
| chr17_-_2690083 | 0.08 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr13_+_31286076 | 0.08 |
ENSDART00000142725
|
gdf10a
|
growth differentiation factor 10a |
| chr19_-_43750108 | 0.08 |
ENSDART00000171806
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr3_+_29714775 | 0.07 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr8_-_13364879 | 0.07 |
ENSDART00000063863
|
ccdc124
|
coiled-coil domain containing 124 |
| chr17_-_23727978 | 0.07 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr1_+_44711446 | 0.06 |
ENSDART00000193481
ENSDART00000003895 |
ssrp1b
|
structure specific recognition protein 1b |
| chr2_+_36112273 | 0.06 |
ENSDART00000191315
|
traj35
|
T-cell receptor alpha joining 35 |
| chr16_-_49505275 | 0.06 |
ENSDART00000160784
|
satb1b
|
SATB homeobox 1b |
| chr3_-_19495814 | 0.06 |
ENSDART00000162248
|
CU571315.3
|
|
| chr5_-_31857345 | 0.05 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr3_+_49097775 | 0.05 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr16_-_10547781 | 0.05 |
ENSDART00000166495
|
g6fl
|
g6f-like |
| chr9_-_31090593 | 0.03 |
ENSDART00000089136
|
gpr18
|
G protein-coupled receptor 18 |
| chr15_+_31344472 | 0.03 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr22_+_2207502 | 0.03 |
ENSDART00000169162
|
si:dkeyp-79b7.12
|
si:dkeyp-79b7.12 |
| chr19_-_617246 | 0.03 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr2_+_19785540 | 0.03 |
ENSDART00000149789
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr17_-_50355512 | 0.03 |
ENSDART00000041685
|
otofb
|
otoferlin b |
| chr24_-_11908115 | 0.03 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr10_+_40660772 | 0.03 |
ENSDART00000148007
|
taar19l
|
trace amine associated receptor 19l |
| chr2_-_43196595 | 0.02 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr7_+_19552381 | 0.02 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr2_-_30734098 | 0.02 |
ENSDART00000133769
|
rp1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr21_-_3770636 | 0.02 |
ENSDART00000053596
|
scamp1
|
secretory carrier membrane protein 1 |
| chr1_+_41588170 | 0.02 |
ENSDART00000139175
|
si:dkey-56e3.2
|
si:dkey-56e3.2 |
| chr13_-_25719628 | 0.02 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr22_+_16308806 | 0.02 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr5_+_45140914 | 0.02 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_19609638 | 0.02 |
ENSDART00000079319
|
rprml
|
reprimo-like |
| chr22_-_30999000 | 0.01 |
ENSDART00000109841
|
ssuh2.3
|
ssu-2 homolog, tandem duplicate 3 |
| chr4_+_19534833 | 0.01 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr6_-_60147517 | 0.01 |
ENSDART00000083453
|
slc32a1
|
solute carrier family 32 (GABA vesicular transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxd11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.0 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.3 | 0.9 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.3 | 0.9 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.2 | 2.6 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 0.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.7 | GO:0071072 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 0.6 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 2.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.6 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 0.6 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.1 | 0.9 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.1 | 1.0 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.5 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.7 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.8 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 1.7 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 1.0 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 0.6 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.3 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 2.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 1.8 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.6 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.7 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.2 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.4 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.3 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 1.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.8 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.4 | GO:0046466 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.3 | 0.9 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 1.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 0.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 1.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.7 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 4.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.4 | 2.6 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 1.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 0.5 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 0.6 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 0.4 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.7 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.1 | 1.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.5 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.4 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.2 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.4 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.9 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 1.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.2 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 1.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 1.7 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.4 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 0.6 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |