Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
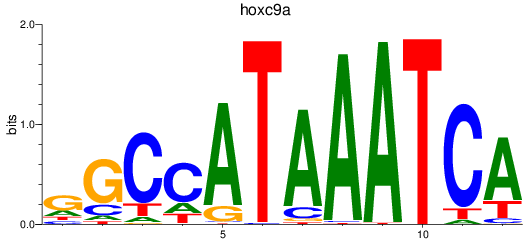
Results for hoxc9a
Z-value: 0.20
Transcription factors associated with hoxc9a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc9a
|
ENSDARG00000092809 | homeobox C9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc9a | dr11_v1_chr23_+_36095260_36095260 | 0.33 | 1.9e-01 | Click! |
Activity profile of hoxc9a motif
Sorted Z-values of hoxc9a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_27830818 | 0.29 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr18_-_39288894 | 0.28 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr1_-_681116 | 0.28 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr6_+_38845697 | 0.27 |
ENSDART00000053187
|
stk35l
|
serine/threonine kinase 35, like |
| chr1_-_6085750 | 0.26 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr3_-_40232615 | 0.24 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr6_+_38626926 | 0.24 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr17_+_19626479 | 0.22 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr6_+_38845848 | 0.22 |
ENSDART00000184907
|
stk35l
|
serine/threonine kinase 35, like |
| chr20_-_53949798 | 0.21 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr25_+_3058924 | 0.20 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr11_-_13341483 | 0.19 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr6_-_43283122 | 0.18 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr11_-_13341051 | 0.18 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr22_+_10713713 | 0.17 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr9_+_44722205 | 0.17 |
ENSDART00000086176
ENSDART00000145271 ENSDART00000132696 |
nckap1
|
NCK-associated protein 1 |
| chr21_-_9769500 | 0.17 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr13_-_22961605 | 0.15 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr2_-_41723487 | 0.15 |
ENSDART00000170171
|
zgc:110158
|
zgc:110158 |
| chr7_+_31051213 | 0.15 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr21_+_22878834 | 0.15 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr2_-_41723165 | 0.14 |
ENSDART00000155577
|
zgc:110158
|
zgc:110158 |
| chr19_+_7424347 | 0.14 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr4_+_10754802 | 0.14 |
ENSDART00000181470
|
stab2
|
stabilin 2 |
| chr10_+_37173029 | 0.13 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr1_-_23157583 | 0.13 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr17_-_25630822 | 0.13 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr22_-_21046843 | 0.13 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr3_-_37699992 | 0.12 |
ENSDART00000151193
|
gpatch8
|
G patch domain containing 8 |
| chr7_+_31051603 | 0.12 |
ENSDART00000108721
|
tjp1a
|
tight junction protein 1a |
| chr12_-_22238004 | 0.11 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr22_-_21046654 | 0.11 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr14_-_14640401 | 0.11 |
ENSDART00000168027
ENSDART00000167521 |
znf185
|
zinc finger protein 185 with LIM domain |
| chr11_+_18873113 | 0.11 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr25_+_15997957 | 0.08 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr14_-_31715373 | 0.08 |
ENSDART00000127303
ENSDART00000173274 ENSDART00000173435 ENSDART00000172876 ENSDART00000173036 |
map7d3
|
MAP7 domain containing 3 |
| chr20_+_23658791 | 0.08 |
ENSDART00000192145
|
si:ch211-191d2.2
|
si:ch211-191d2.2 |
| chr14_-_4044545 | 0.08 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr23_-_3721444 | 0.08 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr17_-_51262430 | 0.08 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr17_+_15788100 | 0.08 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr14_+_35024521 | 0.08 |
ENSDART00000158634
ENSDART00000170631 |
ebf3a
|
early B cell factor 3a |
| chr2_+_34967022 | 0.07 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr4_-_4261673 | 0.07 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr14_-_32959851 | 0.07 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr2_-_18830722 | 0.06 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr5_+_63857055 | 0.06 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr17_-_19626357 | 0.06 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr16_+_37891735 | 0.06 |
ENSDART00000178753
ENSDART00000142104 |
cep162
|
centrosomal protein 162 |
| chr10_+_26571174 | 0.05 |
ENSDART00000148617
ENSDART00000112956 |
slc9a6b
|
solute carrier family 9, subfamily A (NHE6, cation proton antiporter 6), member 6b |
| chr18_-_26510545 | 0.05 |
ENSDART00000135133
|
FRMD5
|
si:ch211-69m14.1 |
| chr9_+_28232522 | 0.05 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr14_+_21783400 | 0.05 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr24_+_39211288 | 0.04 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr3_-_23461954 | 0.04 |
ENSDART00000040065
|
casc3
|
cancer susceptibility candidate 3 |
| chr3_-_8388344 | 0.04 |
ENSDART00000146856
|
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr17_-_20849879 | 0.04 |
ENSDART00000088100
ENSDART00000149630 |
ank3b
|
ankyrin 3b |
| chr9_-_34937025 | 0.04 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr21_+_22878991 | 0.04 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr15_-_14589677 | 0.04 |
ENSDART00000172195
|
coq8b
|
coenzyme Q8B |
| chr8_+_47633438 | 0.04 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr10_+_31953502 | 0.04 |
ENSDART00000185634
|
lhfpl6
|
LHFPL tetraspan subfamily member 6 |
| chr15_+_22722684 | 0.04 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr5_-_51619262 | 0.04 |
ENSDART00000134606
ENSDART00000081249 |
otpb
|
orthopedia homeobox b |
| chr4_-_20235904 | 0.04 |
ENSDART00000146621
ENSDART00000193655 |
stk38l
|
serine/threonine kinase 38 like |
| chr8_+_22931427 | 0.03 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr12_-_33558879 | 0.03 |
ENSDART00000161167
|
mbtd1
|
mbt domain containing 1 |
| chr3_-_51109286 | 0.02 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr8_+_6954984 | 0.02 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr18_-_25855263 | 0.02 |
ENSDART00000042074
|
sema4ba
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ba |
| chr9_-_40014339 | 0.02 |
ENSDART00000166918
|
IKZF2
|
si:zfos-1425h8.1 |
| chr16_-_35952789 | 0.02 |
ENSDART00000180118
|
eva1ba
|
eva-1 homolog Ba (C. elegans) |
| chr25_-_5963535 | 0.02 |
ENSDART00000155751
|
nuak1b
|
NUAK family, SNF1-like kinase, 1b |
| chr8_-_23701880 | 0.02 |
ENSDART00000139897
|
si:ch73-237c6.1
|
si:ch73-237c6.1 |
| chr12_-_33558727 | 0.02 |
ENSDART00000086087
|
mbtd1
|
mbt domain containing 1 |
| chr11_-_40457325 | 0.01 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr4_+_21867522 | 0.01 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr10_-_11385155 | 0.01 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr16_-_16761164 | 0.01 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr5_-_37871526 | 0.01 |
ENSDART00000136450
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr5_-_51619742 | 0.01 |
ENSDART00000188537
|
otpb
|
orthopedia homeobox b |
| chr4_+_16715267 | 0.01 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr7_-_26408472 | 0.00 |
ENSDART00000111494
|
gal3st4
|
galactose-3-O-sulfotransferase 4 |
| chr12_-_6172154 | 0.00 |
ENSDART00000185434
|
a1cf
|
apobec1 complementation factor |
| chr1_-_59422880 | 0.00 |
ENSDART00000167244
|
si:ch211-188p14.2
|
si:ch211-188p14.2 |
| chr25_+_9003879 | 0.00 |
ENSDART00000186228
|
rag1
|
recombination activating gene 1 |
| chr23_+_11374168 | 0.00 |
ENSDART00000163083
|
chl1a
|
cell adhesion molecule L1-like a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc9a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.1 | GO:1905038 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.3 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0005879 | axonemal microtubule(GO:0005879) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |