Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for hoxc8a
Z-value: 0.87
Transcription factors associated with hoxc8a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc8a
|
ENSDARG00000070346 | homeobox C8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc8a | dr11_v1_chr23_+_36101185_36101185 | 0.89 | 6.5e-07 | Click! |
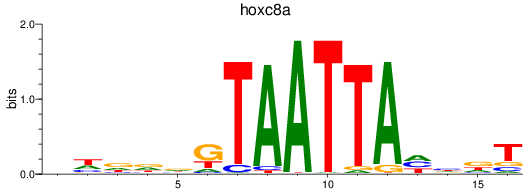
Activity profile of hoxc8a motif
Sorted Z-values of hoxc8a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_51180938 | 3.57 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr17_+_50248152 | 3.24 |
ENSDART00000153998
|
zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr20_-_7072487 | 3.20 |
ENSDART00000145954
|
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr11_-_40728380 | 3.14 |
ENSDART00000023745
|
ccdc114
|
coiled-coil domain containing 114 |
| chr5_-_29671586 | 2.87 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr5_+_29671681 | 2.41 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr2_+_16482338 | 2.30 |
ENSDART00000143912
|
fbxo36b
|
F-box protein 36b |
| chr20_-_14925281 | 1.91 |
ENSDART00000152641
|
dnm3a
|
dynamin 3a |
| chr16_+_4497302 | 1.76 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
| chr20_-_14924858 | 1.69 |
ENSDART00000047039
|
dnm3a
|
dynamin 3a |
| chr1_+_22691256 | 1.47 |
ENSDART00000017413
|
zmynd10
|
zinc finger, MYND-type containing 10 |
| chr4_-_22472653 | 1.43 |
ENSDART00000066903
ENSDART00000130072 ENSDART00000123369 |
kmt2e
|
lysine (K)-specific methyltransferase 2E |
| chrM_+_9735 | 1.39 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr10_+_6884627 | 1.34 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr7_-_40110140 | 1.34 |
ENSDART00000173469
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr5_+_19343880 | 1.31 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr21_+_30355767 | 1.26 |
ENSDART00000189948
|
CR749164.1
|
|
| chr18_+_3037998 | 1.23 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr8_-_13064330 | 1.19 |
ENSDART00000129168
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chrM_+_9052 | 1.05 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr4_-_815871 | 1.05 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr24_-_38657683 | 1.00 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr7_-_45999333 | 0.99 |
ENSDART00000158603
|
si:ch211-260e23.8
|
si:ch211-260e23.8 |
| chr24_+_26997798 | 0.96 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr13_+_13693722 | 0.93 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr8_-_12403077 | 0.92 |
ENSDART00000142150
|
phf19
|
PHD finger protein 19 |
| chr10_+_42358426 | 0.88 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr7_-_48667056 | 0.87 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr3_+_30500968 | 0.87 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr7_-_24046999 | 0.87 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr5_+_69981453 | 0.87 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr10_-_35108683 | 0.87 |
ENSDART00000049633
|
zgc:110006
|
zgc:110006 |
| chr20_+_40457599 | 0.81 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr20_-_45812144 | 0.78 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr25_-_3470910 | 0.71 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr21_-_26490186 | 0.71 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr15_+_15856178 | 0.67 |
ENSDART00000080338
|
dusp14
|
dual specificity phosphatase 14 |
| chr3_+_22442445 | 0.67 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr2_+_35993404 | 0.67 |
ENSDART00000170845
|
lamc2
|
laminin, gamma 2 |
| chr25_+_31868268 | 0.66 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_-_67894046 | 0.66 |
ENSDART00000168964
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr23_+_40604951 | 0.60 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr10_-_5847655 | 0.58 |
ENSDART00000192773
|
ankrd55
|
ankyrin repeat domain 55 |
| chr10_-_26744131 | 0.57 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr21_-_32097908 | 0.54 |
ENSDART00000147387
|
si:ch211-160j14.3
|
si:ch211-160j14.3 |
| chr2_+_20793982 | 0.53 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr8_+_18624658 | 0.53 |
ENSDART00000089141
|
fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr23_-_16485190 | 0.52 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr13_+_35339182 | 0.51 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr2_-_30668580 | 0.49 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr20_+_11731039 | 0.49 |
ENSDART00000152215
ENSDART00000152585 |
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr14_-_30587814 | 0.49 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr16_+_13818743 | 0.47 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr8_+_52637507 | 0.44 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr20_-_9095105 | 0.43 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr6_-_30210378 | 0.42 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr2_-_17235891 | 0.42 |
ENSDART00000144251
|
artnb
|
artemin b |
| chr22_-_1079773 | 0.40 |
ENSDART00000136668
|
si:ch1073-15f12.3
|
si:ch1073-15f12.3 |
| chr10_+_11355841 | 0.38 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr18_+_17827149 | 0.36 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr16_-_28878080 | 0.36 |
ENSDART00000149501
|
si:dkey-239n17.4
|
si:dkey-239n17.4 |
| chr23_+_36130883 | 0.35 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr11_-_1509773 | 0.34 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr22_-_17474583 | 0.33 |
ENSDART00000148027
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr15_-_35252522 | 0.33 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr2_+_4207209 | 0.30 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr15_-_15357178 | 0.30 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr5_+_66433287 | 0.30 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr13_+_31321297 | 0.28 |
ENSDART00000143308
|
antxr1d
|
anthrax toxin receptor 1d |
| chr2_+_31833997 | 0.28 |
ENSDART00000066788
|
epdr1
|
ependymin related 1 |
| chr11_-_21528056 | 0.27 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr4_+_14727018 | 0.27 |
ENSDART00000124189
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr11_+_33818179 | 0.25 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr9_-_21918963 | 0.24 |
ENSDART00000090782
|
lmo7a
|
LIM domain 7a |
| chr16_-_27749172 | 0.24 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr4_+_14727212 | 0.24 |
ENSDART00000158094
|
cmasa
|
cytidine monophosphate N-acetylneuraminic acid synthetase a |
| chr7_+_48667081 | 0.23 |
ENSDART00000083473
|
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr6_-_43283122 | 0.23 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr13_+_36622100 | 0.23 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr1_-_40911332 | 0.23 |
ENSDART00000027463
|
hmx4
|
H6 family homeobox 4 |
| chr22_-_17474781 | 0.21 |
ENSDART00000186817
|
si:ch211-197g15.8
|
si:ch211-197g15.8 |
| chr21_+_22738939 | 0.21 |
ENSDART00000151342
ENSDART00000079145 |
arhgap42a
|
Rho GTPase activating protein 42a |
| chr10_+_2899108 | 0.19 |
ENSDART00000147031
|
erap1a
|
endoplasmic reticulum aminopeptidase 1a |
| chr6_+_18321627 | 0.18 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr21_+_29077509 | 0.18 |
ENSDART00000128561
|
ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr11_-_6188413 | 0.17 |
ENSDART00000109972
|
ccl44
|
chemokine (C-C motif) ligand 44 |
| chr5_-_42904329 | 0.17 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr22_+_1170294 | 0.16 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr15_-_42736433 | 0.15 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr4_-_5333971 | 0.15 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr13_-_31812394 | 0.14 |
ENSDART00000124445
|
sertad4
|
SERTA domain containing 4 |
| chr11_+_30244356 | 0.14 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr17_-_43556208 | 0.14 |
ENSDART00000188424
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr19_+_16064439 | 0.13 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr21_-_35419486 | 0.13 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr18_+_27738349 | 0.11 |
ENSDART00000187816
|
tspan18b
|
tetraspanin 18b |
| chr10_-_21955231 | 0.11 |
ENSDART00000183695
|
FO744833.2
|
|
| chr1_-_58036509 | 0.11 |
ENSDART00000081122
|
COLGALT1
|
si:ch211-114l13.7 |
| chr5_+_63857055 | 0.10 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr11_-_11266882 | 0.09 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr13_+_12299997 | 0.09 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr15_-_14552101 | 0.08 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_-_43556415 | 0.08 |
ENSDART00000190102
ENSDART00000193156 |
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr20_-_9462433 | 0.07 |
ENSDART00000152674
ENSDART00000040557 |
zgc:101840
|
zgc:101840 |
| chr11_+_11267493 | 0.07 |
ENSDART00000148425
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr8_-_31107537 | 0.07 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr8_+_22516728 | 0.07 |
ENSDART00000146013
|
si:ch211-261n11.3
|
si:ch211-261n11.3 |
| chr17_+_26611929 | 0.07 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr8_-_30204650 | 0.07 |
ENSDART00000133209
|
zgc:162939
|
zgc:162939 |
| chr12_-_4532066 | 0.06 |
ENSDART00000092687
|
trpm4b.2
|
transient receptor potential cation channel, subfamily M, member 4b, transient receptor potential cation channel, subfamily M, member 4b, tandem duplicate 2 |
| chr15_+_31899312 | 0.06 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr6_+_40591149 | 0.06 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr14_-_46173265 | 0.05 |
ENSDART00000164321
|
zmp:0000000758
|
zmp:0000000758 |
| chr25_-_13490744 | 0.05 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr1_+_53842344 | 0.05 |
ENSDART00000114506
|
crfb17
|
cytokine receptor family member B17 |
| chr15_-_43284021 | 0.05 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr15_+_841383 | 0.04 |
ENSDART00000115077
|
zgc:174573
|
zgc:174573 |
| chr19_+_10592778 | 0.04 |
ENSDART00000135488
ENSDART00000151624 |
si:dkey-211g8.5
|
si:dkey-211g8.5 |
| chr6_+_24398907 | 0.04 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr2_+_3502430 | 0.04 |
ENSDART00000113395
|
SERP1
|
zgc:92744 |
| chr14_-_27121854 | 0.04 |
ENSDART00000173119
|
pcdh11
|
protocadherin 11 |
| chr11_+_37250839 | 0.04 |
ENSDART00000170209
|
il17rc
|
interleukin 17 receptor C |
| chr13_+_31070181 | 0.03 |
ENSDART00000110560
ENSDART00000146088 |
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr23_-_10831995 | 0.03 |
ENSDART00000142533
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr21_-_5856050 | 0.03 |
ENSDART00000115367
|
CABZ01071020.1
|
|
| chr4_-_49582758 | 0.03 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr10_+_40568735 | 0.03 |
ENSDART00000136468
ENSDART00000182841 |
taar18i
|
trace amine associated receptor 18i |
| chr16_-_21038015 | 0.02 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr11_-_7380674 | 0.01 |
ENSDART00000014979
ENSDART00000103418 |
vtg3
|
vitellogenin 3, phosvitinless |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc8a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.4 | 1.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 1.3 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 3.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.2 | 0.9 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.1 | 0.7 | GO:0003096 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.9 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.9 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 1.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.5 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.3 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.1 | 1.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.3 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 0.5 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 3.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.0 | 0.8 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.6 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.2 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.2 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.0 | 0.7 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 3.7 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.2 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 3.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.2 | 1.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.1 | 1.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.1 | 0.7 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 1.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 1.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.3 | 1.3 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 3.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 1.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 3.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.7 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |