Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
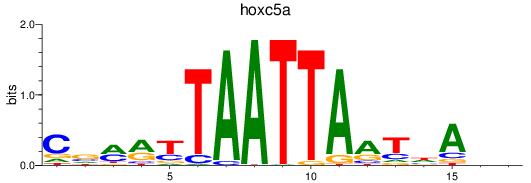
Results for hoxc5a
Z-value: 0.60
Transcription factors associated with hoxc5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc5a
|
ENSDARG00000070340 | homeobox C5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc5a | dr11_v1_chr23_+_36118738_36118738 | 0.39 | 1.1e-01 | Click! |
Activity profile of hoxc5a motif
Sorted Z-values of hoxc5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_20939579 | 2.23 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr4_+_17279966 | 1.78 |
ENSDART00000067005
ENSDART00000137487 |
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr17_+_15433518 | 1.52 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr18_+_46382484 | 1.50 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr17_+_15433671 | 1.35 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr5_+_69747417 | 1.34 |
ENSDART00000153717
|
si:ch211-275j6.5
|
si:ch211-275j6.5 |
| chr17_+_41992054 | 1.32 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr11_-_2838699 | 1.29 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr17_-_39772999 | 1.27 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr15_-_46779934 | 1.24 |
ENSDART00000085136
|
clcn2c
|
chloride channel 2c |
| chr2_+_49799470 | 1.24 |
ENSDART00000146325
|
si:ch211-190k17.19
|
si:ch211-190k17.19 |
| chr2_-_38000276 | 1.20 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr20_-_51355465 | 1.19 |
ENSDART00000151620
ENSDART00000151690 ENSDART00000110289 |
tcte1
|
t-complex-associated-testis-expressed 1 |
| chr19_+_43297546 | 1.19 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr18_+_7456597 | 1.18 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr5_+_42400777 | 1.17 |
ENSDART00000183114
|
BX548073.8
|
|
| chr5_+_38040407 | 1.17 |
ENSDART00000139936
|
si:dkey-111e8.4
|
si:dkey-111e8.4 |
| chr9_-_41062412 | 1.14 |
ENSDART00000193879
|
ankar
|
ankyrin and armadillo repeat containing |
| chr8_+_25569903 | 1.12 |
ENSDART00000062375
|
si:dkey-48j7.3
|
si:dkey-48j7.3 |
| chr25_+_37293312 | 1.10 |
ENSDART00000086737
ENSDART00000161595 |
si:dkey-234i14.9
|
si:dkey-234i14.9 |
| chr21_+_45502621 | 1.08 |
ENSDART00000166719
|
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr8_-_13046089 | 1.05 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr24_+_12989727 | 1.05 |
ENSDART00000126842
ENSDART00000129309 |
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr3_+_26145013 | 1.05 |
ENSDART00000162546
ENSDART00000129561 |
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr14_-_858985 | 1.04 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr1_+_49668423 | 1.04 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr18_+_7456888 | 1.02 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr9_+_7732714 | 1.02 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr21_+_11885404 | 1.01 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr3_+_60721342 | 1.00 |
ENSDART00000157772
|
foxj1a
|
forkhead box J1a |
| chr22_-_36530902 | 0.99 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr10_-_31015535 | 0.98 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr23_+_12134839 | 0.97 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr5_-_54672763 | 0.96 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr20_-_13640598 | 0.95 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr8_-_44586981 | 0.95 |
ENSDART00000026831
ENSDART00000113945 |
rsph14
|
radial spoke head 14 homolog |
| chr4_+_9669717 | 0.95 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr5_-_71705191 | 0.92 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr14_+_17397534 | 0.90 |
ENSDART00000180162
ENSDART00000129838 |
rnf212
|
ring finger protein 212 |
| chr23_-_19140781 | 0.89 |
ENSDART00000143580
|
si:ch73-381f5.2
|
si:ch73-381f5.2 |
| chr6_+_6797520 | 0.88 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr20_+_21022641 | 0.88 |
ENSDART00000152561
ENSDART00000141591 |
si:dkey-219e20.2
|
si:dkey-219e20.2 |
| chr9_+_23895711 | 0.87 |
ENSDART00000034686
|
cops8
|
COP9 signalosome subunit 8 |
| chr19_+_31873308 | 0.86 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr14_+_46313396 | 0.86 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr21_+_45502773 | 0.84 |
ENSDART00000160059
ENSDART00000165704 |
si:dkey-223p19.2
|
si:dkey-223p19.2 |
| chr9_-_30576522 | 0.83 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr10_-_43771447 | 0.83 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr9_-_24970018 | 0.83 |
ENSDART00000026924
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr2_+_44571200 | 0.82 |
ENSDART00000098132
|
klhl24a
|
kelch-like family member 24a |
| chr15_-_45110011 | 0.81 |
ENSDART00000182047
ENSDART00000188662 |
CABZ01072607.1
|
|
| chr19_+_43969363 | 0.80 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr6_+_49551614 | 0.80 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr20_+_25586099 | 0.79 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr15_-_45246911 | 0.79 |
ENSDART00000189557
ENSDART00000185291 |
CABZ01072607.1
|
|
| chr15_-_18162647 | 0.79 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr16_-_42894628 | 0.79 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr21_+_18292535 | 0.79 |
ENSDART00000170205
ENSDART00000169676 ENSDART00000163063 |
dnai1.1
|
dynein, axonemal, intermediate chain 1, paralog 1 |
| chr19_+_40177246 | 0.78 |
ENSDART00000049147
|
rbm48
|
RNA binding motif protein 48 |
| chr1_-_19502322 | 0.78 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr21_-_32301109 | 0.77 |
ENSDART00000139890
|
clk4b
|
CDC-like kinase 4b |
| chr12_-_35944654 | 0.77 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr23_+_39695827 | 0.76 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr3_-_39180048 | 0.76 |
ENSDART00000049720
|
cdk21
|
cyclin-dependent kinase 21 |
| chr6_-_40842768 | 0.75 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr5_+_42386705 | 0.73 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 0.73 |
ENSDART00000188489
|
BX548073.11
|
|
| chr22_-_26865361 | 0.73 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr1_+_33697170 | 0.72 |
ENSDART00000131664
|
NSUN3
|
NOP2/Sun RNA methyltransferase family member 3 |
| chr7_+_49681040 | 0.72 |
ENSDART00000176372
ENSDART00000192172 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr5_+_42393896 | 0.72 |
ENSDART00000189550
|
BX548073.13
|
|
| chr6_-_7720332 | 0.72 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr15_-_39955785 | 0.72 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr12_+_47698356 | 0.72 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr20_+_19512727 | 0.71 |
ENSDART00000063696
|
atraid
|
all-trans retinoic acid-induced differentiation factor |
| chr16_-_28856112 | 0.71 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_+_42379517 | 0.70 |
ENSDART00000103325
|
pimr59
|
Pim proto-oncogene, serine/threonine kinase, related 59 |
| chr20_+_42881104 | 0.70 |
ENSDART00000131338
|
pimr110
|
Pim proto-oncogene, serine/threonine kinase, related 110 |
| chr23_-_42752550 | 0.70 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr7_+_23515966 | 0.70 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr21_-_43015383 | 0.70 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr5_-_42661012 | 0.70 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr18_-_48547564 | 0.70 |
ENSDART00000138607
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr7_+_73397283 | 0.69 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr2_-_1548330 | 0.68 |
ENSDART00000082155
ENSDART00000108481 ENSDART00000111272 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr15_+_22267847 | 0.67 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr8_-_11546175 | 0.67 |
ENSDART00000081909
|
si:ch211-248e11.2
|
si:ch211-248e11.2 |
| chr20_+_7584211 | 0.67 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr14_-_49063157 | 0.66 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr3_+_62196672 | 0.66 |
ENSDART00000097312
|
sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr1_+_55239710 | 0.66 |
ENSDART00000174846
|
si:ch211-286b5.2
|
si:ch211-286b5.2 |
| chr23_-_42752387 | 0.65 |
ENSDART00000149781
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr7_-_52388734 | 0.65 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr10_+_40324395 | 0.63 |
ENSDART00000147205
|
gltpb
|
glycolipid transfer protein b |
| chr24_-_37955993 | 0.63 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr7_-_4461104 | 0.63 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr12_+_7497882 | 0.62 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr17_+_6625717 | 0.62 |
ENSDART00000190753
ENSDART00000181298 |
BX005321.1
|
|
| chr23_-_23401305 | 0.62 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr21_-_2299002 | 0.62 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr5_-_41494831 | 0.62 |
ENSDART00000051081
|
eef2l2
|
eukaryotic translation elongation factor 2, like 2 |
| chr12_-_35830625 | 0.61 |
ENSDART00000180028
|
CU459056.1
|
|
| chr9_-_7652792 | 0.61 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr25_+_20715950 | 0.60 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr2_+_33751490 | 0.59 |
ENSDART00000145031
|
si:dkey-31m5.7
|
si:dkey-31m5.7 |
| chr8_+_31435452 | 0.59 |
ENSDART00000145282
|
selenop
|
selenoprotein P |
| chr13_+_4409294 | 0.59 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr21_-_25801956 | 0.59 |
ENSDART00000101219
|
mettl27
|
methyltransferase like 27 |
| chr1_-_669717 | 0.58 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr11_+_30057762 | 0.58 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_+_36284986 | 0.58 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr12_+_45200744 | 0.58 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr9_-_9415000 | 0.57 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr3_-_13461056 | 0.57 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr13_-_37620091 | 0.56 |
ENSDART00000135875
ENSDART00000193270 ENSDART00000018064 |
zgc:152791
|
zgc:152791 |
| chr4_+_22480169 | 0.56 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr16_+_54780544 | 0.56 |
ENSDART00000126646
|
si:zfos-1192g2.3
|
si:zfos-1192g2.3 |
| chr1_-_5394752 | 0.56 |
ENSDART00000103724
ENSDART00000188453 |
ndfip2
|
Nedd4 family interacting protein 2 |
| chr16_+_10972733 | 0.56 |
ENSDART00000049323
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr10_+_26747755 | 0.56 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr2_+_19522082 | 0.56 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr15_-_28200049 | 0.55 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr2_-_56348727 | 0.55 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr21_-_8422351 | 0.55 |
ENSDART00000055329
ENSDART00000134360 |
lhx2a
|
LIM homeobox 2a |
| chr1_-_54063520 | 0.55 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr24_+_35787629 | 0.54 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr5_+_44944778 | 0.54 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr17_+_51682429 | 0.54 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr9_+_18716485 | 0.53 |
ENSDART00000135125
|
serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr19_-_1855368 | 0.53 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr1_-_22851481 | 0.53 |
ENSDART00000054386
|
qdprb1
|
quinoid dihydropteridine reductase b1 |
| chr5_+_2815021 | 0.53 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr6_+_52350443 | 0.53 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr9_-_21382190 | 0.52 |
ENSDART00000110943
ENSDART00000188129 ENSDART00000193876 |
ift88
|
intraflagellar transport 88 homolog |
| chr3_+_58833306 | 0.52 |
ENSDART00000113223
|
igl1c3
|
immunoglobulin light 1 constant 3 |
| chr3_-_40658820 | 0.52 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr19_-_3931917 | 0.52 |
ENSDART00000162532
|
map7d1b
|
MAP7 domain containing 1b |
| chr22_+_34663328 | 0.51 |
ENSDART00000183912
|
si:ch73-304f21.1
|
si:ch73-304f21.1 |
| chr1_-_55785722 | 0.51 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr1_-_19845378 | 0.51 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr2_+_38373272 | 0.50 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr10_+_6625785 | 0.50 |
ENSDART00000148433
|
si:ch211-57m13.3
|
si:ch211-57m13.3 |
| chr21_-_18972206 | 0.50 |
ENSDART00000146743
|
pimr72
|
Pim proto-oncogene, serine/threonine kinase, related 72 |
| chr20_+_5985329 | 0.50 |
ENSDART00000165489
|
cep128
|
centrosomal protein 128 |
| chr22_-_26865181 | 0.50 |
ENSDART00000138311
|
hmox2a
|
heme oxygenase 2a |
| chr16_+_28728347 | 0.49 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr13_-_9450210 | 0.49 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr3_-_15999501 | 0.49 |
ENSDART00000160668
|
nme3
|
NME/NM23 nucleoside diphosphate kinase 3 |
| chr14_-_24761132 | 0.49 |
ENSDART00000146299
|
slit3
|
slit homolog 3 (Drosophila) |
| chr8_-_17167819 | 0.48 |
ENSDART00000135042
ENSDART00000143920 |
mrps36
|
mitochondrial ribosomal protein S36 |
| chr22_+_508290 | 0.48 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr5_-_9073433 | 0.48 |
ENSDART00000099891
|
atp5meb
|
ATP synthase membrane subunit eb |
| chr14_+_39258569 | 0.48 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr5_-_13251907 | 0.48 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr14_-_6225336 | 0.48 |
ENSDART00000111681
|
hdx
|
highly divergent homeobox |
| chr6_-_9922266 | 0.47 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr10_-_9410068 | 0.47 |
ENSDART00000041382
|
morn5
|
MORN repeat containing 5 |
| chr10_-_34046757 | 0.47 |
ENSDART00000099648
|
pimr149
|
Pim proto-oncogene, serine/threonine kinase, related 149 |
| chr6_-_54179860 | 0.47 |
ENSDART00000164283
|
rps10
|
ribosomal protein S10 |
| chr4_+_57881965 | 0.47 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr1_+_54037077 | 0.46 |
ENSDART00000109386
|
triobpa
|
TRIO and F-actin binding protein a |
| chr21_+_45841731 | 0.46 |
ENSDART00000038657
|
faxdc2
|
fatty acid hydroxylase domain containing 2 |
| chr5_-_1203455 | 0.46 |
ENSDART00000172177
|
surf4
|
surfeit gene 4 |
| chr8_-_45760087 | 0.46 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr18_-_48983690 | 0.46 |
ENSDART00000182359
|
FO681288.3
|
|
| chr24_+_32176155 | 0.45 |
ENSDART00000003745
|
vim
|
vimentin |
| chr9_-_47472998 | 0.45 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr4_+_22297839 | 0.45 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr11_-_25733910 | 0.45 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr25_+_21833287 | 0.45 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr9_-_30555725 | 0.45 |
ENSDART00000079222
|
chaf1b
|
chromatin assembly factor 1, subunit B |
| chr1_+_59293873 | 0.45 |
ENSDART00000168036
|
rdh8b
|
retinol dehydrogenase 8b |
| chr2_+_9990491 | 0.45 |
ENSDART00000011906
|
slc35a3b
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3b |
| chr7_+_26534131 | 0.44 |
ENSDART00000173980
|
si:dkey-62k3.5
|
si:dkey-62k3.5 |
| chr3_-_53114299 | 0.44 |
ENSDART00000109390
|
AL954361.1
|
|
| chr14_-_413273 | 0.44 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr21_+_22840246 | 0.44 |
ENSDART00000151621
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr14_-_7137808 | 0.44 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr2_+_33726862 | 0.43 |
ENSDART00000146745
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr6_-_31361546 | 0.43 |
ENSDART00000027550
|
ak4
|
adenylate kinase 4 |
| chr21_+_45223194 | 0.43 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr6_+_30091811 | 0.43 |
ENSDART00000088403
|
meltf
|
melanotransferrin |
| chr1_-_53259746 | 0.43 |
ENSDART00000134222
|
il15
|
interleukin 15 |
| chr16_+_46205693 | 0.43 |
ENSDART00000058385
ENSDART00000168145 |
bola1
|
bolA family member 1 |
| chr24_-_38657683 | 0.43 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr3_-_12026741 | 0.43 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr14_-_28001986 | 0.43 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr4_-_9909371 | 0.42 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr22_+_19366866 | 0.42 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr16_-_41004731 | 0.42 |
ENSDART00000102591
|
KCNV1
|
si:dkey-201i6.2 |
| chr21_-_13972745 | 0.42 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr14_+_15257658 | 0.42 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr21_-_19314618 | 0.42 |
ENSDART00000188744
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr6_+_9427641 | 0.42 |
ENSDART00000022620
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr11_-_1509773 | 0.42 |
ENSDART00000050762
|
phactr3b
|
phosphatase and actin regulator 3b |
| chr16_-_52916248 | 0.42 |
ENSDART00000113931
|
zdhhc11
|
zinc finger, DHHC-type containing 11 |
| chr10_-_34123733 | 0.42 |
ENSDART00000157708
ENSDART00000143954 |
pimr150
|
Pim proto-oncogene, serine/threonine kinase, related 150 |
| chr21_-_22827548 | 0.42 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr3_-_40896927 | 0.41 |
ENSDART00000155279
|
ap5z1
|
adaptor-related protein complex 5, zeta 1 subunit |
| chr16_+_23303859 | 0.41 |
ENSDART00000006093
|
slc50a1
|
solute carrier family 50 (sugar efflux transporter), member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0034397 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.4 | 1.8 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.4 | 1.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 0.9 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.3 | 1.1 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 1.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.2 | 1.0 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 0.8 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.2 | 1.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 0.6 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.2 | 0.6 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.5 | GO:0030238 | male sex determination(GO:0030238) |
| 0.2 | 0.7 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.2 | 0.5 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 | 1.0 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.0 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 1.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.1 | 3.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.4 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 0.4 | GO:0060547 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 1.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 0.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.1 | 0.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.3 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.1 | 0.3 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.5 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.6 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.7 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.2 | GO:0071635 | transforming growth factor beta activation(GO:0036363) regulation of transforming growth factor beta production(GO:0071634) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 | 0.7 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) negative regulation of mesoderm development(GO:2000381) |
| 0.1 | 0.4 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.1 | 0.2 | GO:0045601 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 1.3 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.4 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.8 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.5 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.6 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.1 | 0.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.2 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) adenohypophysis morphogenesis(GO:0048855) |
| 0.1 | 0.4 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.3 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 0.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.7 | GO:0051255 | spindle midzone assembly(GO:0051255) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 1.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.3 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.3 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.0 | 0.4 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.8 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.0 | 0.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.5 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.9 | GO:0007129 | synapsis(GO:0007129) |
| 0.0 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0071632 | optomotor response(GO:0071632) |
| 0.0 | 11.9 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.3 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.3 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.0 | 0.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.7 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.2 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:0000972 | transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000972) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.6 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.1 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.3 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0016074 | snoRNA metabolic process(GO:0016074) |
| 0.0 | 0.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.8 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.1 | GO:1901017 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0034205 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0070445 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.8 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0050871 | positive regulation of B cell activation(GO:0050871) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.4 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.1 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.0 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.4 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0046144 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.3 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.1 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.5 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 1.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.1 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.4 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 0.6 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 0.5 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 2.2 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.7 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.4 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.6 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.0 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 0.6 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.3 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.7 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 1.0 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 0.2 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 1.1 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 1.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.7 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.4 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.2 | 2.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.5 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.2 | 1.0 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 0.5 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.8 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.6 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 1.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.6 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.2 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.1 | 0.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.3 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.3 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.6 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.0 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.5 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 1.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.7 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.2 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.0 | 0.3 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0015157 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.5 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.6 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.4 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.3 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0070915 | bioactive lipid receptor activity(GO:0045125) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.0 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.1 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |