Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for hoxc4a
Z-value: 0.38
Transcription factors associated with hoxc4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc4a
|
ENSDARG00000070338 | homeobox C4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc4a | dr11_v1_chr23_+_36130883_36130883 | 0.64 | 3.9e-03 | Click! |
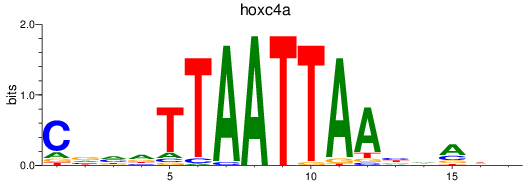
Activity profile of hoxc4a motif
Sorted Z-values of hoxc4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_7456597 | 0.84 |
ENSDART00000142270
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr10_-_29236860 | 0.74 |
ENSDART00000111620
|
ccdc83
|
coiled-coil domain containing 83 |
| chr18_+_7456888 | 0.72 |
ENSDART00000081468
|
terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr11_-_2838699 | 0.66 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr16_-_42894628 | 0.64 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr14_-_858985 | 0.63 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr17_+_41992054 | 0.60 |
ENSDART00000182878
ENSDART00000111537 |
kiz
|
kizuna centrosomal protein |
| chr10_-_31015535 | 0.56 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr16_+_23431189 | 0.48 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr12_-_35830625 | 0.46 |
ENSDART00000180028
|
CU459056.1
|
|
| chr7_-_52388734 | 0.45 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr7_+_23515966 | 0.45 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr12_+_7497882 | 0.39 |
ENSDART00000134608
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr1_-_669717 | 0.36 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr23_+_12134839 | 0.36 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr22_+_508290 | 0.34 |
ENSDART00000135403
|
nuak2
|
NUAK family, SNF1-like kinase, 2 |
| chr8_-_53044300 | 0.31 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr19_+_43297546 | 0.30 |
ENSDART00000168002
|
laptm5
|
lysosomal protein transmembrane 5 |
| chr10_+_26747755 | 0.29 |
ENSDART00000100329
|
f9b
|
coagulation factor IXb |
| chr22_-_16042243 | 0.28 |
ENSDART00000062633
|
s1pr1
|
sphingosine-1-phosphate receptor 1 |
| chr4_+_57881965 | 0.28 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr1_+_16625678 | 0.27 |
ENSDART00000164899
|
pcm1
|
pericentriolar material 1 |
| chr17_-_26604946 | 0.27 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr12_+_17620067 | 0.27 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr9_-_47472998 | 0.26 |
ENSDART00000134480
|
tns1b
|
tensin 1b |
| chr23_+_7471072 | 0.26 |
ENSDART00000135551
|
si:ch211-200e2.1
|
si:ch211-200e2.1 |
| chr4_+_9669717 | 0.26 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr13_-_9450210 | 0.25 |
ENSDART00000133768
|
pimr152
|
Pim proto-oncogene, serine/threonine kinase, related 152 |
| chr10_-_14556978 | 0.25 |
ENSDART00000126643
|
zgc:153395
|
zgc:153395 |
| chr24_-_38657683 | 0.24 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr16_+_40024883 | 0.24 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr20_+_40457599 | 0.24 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr24_+_744713 | 0.23 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr3_-_16039619 | 0.23 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr4_+_16715267 | 0.23 |
ENSDART00000143849
|
pkp2
|
plakophilin 2 |
| chr2_+_20793982 | 0.23 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr16_-_12173554 | 0.23 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr9_-_32177117 | 0.22 |
ENSDART00000078568
|
sf3b1
|
splicing factor 3b, subunit 1 |
| chr20_-_9095105 | 0.22 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr5_+_11840905 | 0.22 |
ENSDART00000030444
|
tesca
|
tescalcin a |
| chr1_-_19502322 | 0.22 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr19_+_31873308 | 0.22 |
ENSDART00000146560
ENSDART00000133045 |
si:dkeyp-34f6.4
|
si:dkeyp-34f6.4 |
| chr6_+_25257728 | 0.21 |
ENSDART00000162581
|
kyat3
|
kynurenine aminotransferase 3 |
| chr13_+_646700 | 0.21 |
ENSDART00000006892
|
tp53bp2a
|
tumor protein p53 binding protein, 2a |
| chr15_+_32798333 | 0.21 |
ENSDART00000162370
ENSDART00000166525 |
spartb
|
spartin b |
| chr19_+_46158078 | 0.21 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr2_-_30668580 | 0.20 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr4_-_9891874 | 0.20 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr4_+_45148652 | 0.20 |
ENSDART00000150798
|
si:dkey-51d8.9
|
si:dkey-51d8.9 |
| chr22_-_12160283 | 0.20 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr7_+_38811800 | 0.20 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr1_+_51039558 | 0.19 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr16_-_12173399 | 0.18 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr8_+_30664077 | 0.18 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr21_-_13123176 | 0.18 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr23_-_16485190 | 0.18 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr21_+_25802190 | 0.17 |
ENSDART00000128987
|
nf2b
|
neurofibromin 2b (merlin) |
| chr10_+_44956660 | 0.17 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr9_-_30576522 | 0.17 |
ENSDART00000101085
|
morc3a
|
MORC family CW-type zinc finger 3a |
| chr15_-_43284021 | 0.16 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr11_+_24815667 | 0.16 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr16_+_54209504 | 0.16 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr9_-_22834860 | 0.16 |
ENSDART00000146486
|
neb
|
nebulin |
| chr11_+_34760628 | 0.16 |
ENSDART00000087216
|
si:dkey-202e22.2
|
si:dkey-202e22.2 |
| chr19_+_43359075 | 0.16 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr23_+_20669149 | 0.16 |
ENSDART00000138936
|
arhgef3l
|
Rho guanine nucleotide exchange factor (GEF) 3, like |
| chr5_-_21422390 | 0.16 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr19_-_8877469 | 0.15 |
ENSDART00000193951
|
celf3a
|
cugbp, Elav-like family member 3a |
| chr18_+_2228737 | 0.15 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr21_+_25236297 | 0.15 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr24_+_28953089 | 0.15 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr6_+_11249706 | 0.15 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr18_+_3037998 | 0.14 |
ENSDART00000185844
ENSDART00000162657 |
rps3
|
ribosomal protein S3 |
| chr4_-_2219705 | 0.14 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr6_+_15373153 | 0.14 |
ENSDART00000155865
|
tmtops2a
|
teleost multiple tissue opsin 2a |
| chr12_+_20641102 | 0.14 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr18_+_13162728 | 0.14 |
ENSDART00000101472
|
tat
|
tyrosine aminotransferase |
| chr24_+_13316737 | 0.14 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr16_+_46111849 | 0.14 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr3_-_29941357 | 0.14 |
ENSDART00000147732
ENSDART00000137973 ENSDART00000103523 |
grna
|
granulin a |
| chr25_-_13320986 | 0.14 |
ENSDART00000169238
|
si:ch211-194m7.8
|
si:ch211-194m7.8 |
| chr9_-_14504834 | 0.14 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr3_+_27606505 | 0.13 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr19_+_11855330 | 0.13 |
ENSDART00000145380
|
adck5
|
aarF domain containing kinase 5 |
| chr3_-_40658820 | 0.13 |
ENSDART00000191948
|
rnf216
|
ring finger protein 216 |
| chr11_+_30057762 | 0.12 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr4_+_77943184 | 0.11 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_+_11250033 | 0.11 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr1_-_45616470 | 0.11 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_+_36284986 | 0.11 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr10_-_11261565 | 0.10 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr11_+_45436703 | 0.10 |
ENSDART00000168295
ENSDART00000173293 |
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr6_-_43283122 | 0.10 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr10_+_16225117 | 0.10 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr18_+_17827149 | 0.10 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr3_-_48716422 | 0.10 |
ENSDART00000164979
|
si:ch211-114m9.1
|
si:ch211-114m9.1 |
| chr24_+_7800486 | 0.09 |
ENSDART00000145504
|
ptprh
|
protein tyrosine phosphatase, receptor type, h |
| chr14_-_34771371 | 0.09 |
ENSDART00000160598
ENSDART00000150413 ENSDART00000168910 |
ablim3
|
actin binding LIM protein family, member 3 |
| chr7_+_34620418 | 0.08 |
ENSDART00000081338
|
slc9a5
|
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr23_+_4709607 | 0.08 |
ENSDART00000166503
ENSDART00000158752 ENSDART00000163860 ENSDART00000172739 |
raf1a
raf1a
|
Raf-1 proto-oncogene, serine/threonine kinase a Raf-1 proto-oncogene, serine/threonine kinase a |
| chr15_+_28318005 | 0.08 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr14_-_413273 | 0.08 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr18_-_17485419 | 0.08 |
ENSDART00000018764
|
foxl1
|
forkhead box L1 |
| chr21_+_8276989 | 0.07 |
ENSDART00000028265
|
nr5a1b
|
nuclear receptor subfamily 5, group A, member 1b |
| chr25_-_21031007 | 0.07 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr15_+_5360407 | 0.06 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr16_-_42965192 | 0.06 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr6_-_43677125 | 0.06 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr10_+_42423318 | 0.06 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr15_+_22867174 | 0.06 |
ENSDART00000035812
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr12_+_48480632 | 0.06 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr8_-_45760087 | 0.06 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr6_-_8392104 | 0.06 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr16_+_12836143 | 0.06 |
ENSDART00000067741
|
cacng6b
|
calcium channel, voltage-dependent, gamma subunit 6b |
| chr22_-_10156581 | 0.05 |
ENSDART00000168304
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr13_+_29462249 | 0.05 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr22_+_12770877 | 0.05 |
ENSDART00000044683
|
ftcd
|
formimidoyltransferase cyclodeaminase |
| chr2_+_47708853 | 0.05 |
ENSDART00000124307
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr2_+_36701322 | 0.05 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr17_-_12336987 | 0.05 |
ENSDART00000172001
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr22_+_19552987 | 0.05 |
ENSDART00000105315
|
hsd11b1la
|
hydroxysteroid (11-beta) dehydrogenase 1-like a |
| chr6_+_24420523 | 0.05 |
ENSDART00000185461
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr10_+_35358675 | 0.05 |
ENSDART00000193263
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr1_+_418869 | 0.05 |
ENSDART00000152173
|
tpp2
|
tripeptidyl peptidase 2 |
| chr1_+_49668423 | 0.05 |
ENSDART00000150880
|
tsga10
|
testis specific, 10 |
| chr23_-_17657348 | 0.05 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr1_+_21731382 | 0.05 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr4_-_13548806 | 0.04 |
ENSDART00000067155
|
il22
|
interleukin 22 |
| chr4_+_36150332 | 0.04 |
ENSDART00000161697
|
znf1116
|
zinc finger protein 1116 |
| chr23_-_1660708 | 0.04 |
ENSDART00000175138
|
CU693481.1
|
|
| chr19_+_2631565 | 0.04 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr20_+_34717403 | 0.04 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr12_-_6880694 | 0.04 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr22_-_19552796 | 0.04 |
ENSDART00000148088
ENSDART00000105485 |
si:dkey-78l4.14
|
si:dkey-78l4.14 |
| chr1_+_6172786 | 0.04 |
ENSDART00000126468
|
prkag3a
|
protein kinase, AMP-activated, gamma 3a non-catalytic subunit |
| chr13_-_52089003 | 0.04 |
ENSDART00000187600
|
tmem254
|
transmembrane protein 254 |
| chr15_+_31899312 | 0.04 |
ENSDART00000155315
|
frya
|
furry homolog a (Drosophila) |
| chr24_-_25691020 | 0.04 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr16_-_31622777 | 0.03 |
ENSDART00000137311
ENSDART00000002930 |
phf20l1
|
PHD finger protein 20 like 1 |
| chr22_+_34784075 | 0.03 |
ENSDART00000167538
|
lcor
|
ligand dependent nuclear receptor corepressor |
| chr2_+_2223837 | 0.03 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr22_+_23359369 | 0.03 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr2_+_38373272 | 0.03 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr8_+_7801060 | 0.03 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr2_-_37140423 | 0.03 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr25_+_13321307 | 0.03 |
ENSDART00000161284
|
si:ch211-194m7.5
|
si:ch211-194m7.5 |
| chr17_+_2162916 | 0.03 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr17_+_50524573 | 0.02 |
ENSDART00000187974
|
CR382385.1
|
|
| chr17_-_15229787 | 0.02 |
ENSDART00000039165
|
styx
|
serine/threonine/tyrosine interacting protein |
| chr11_-_45141309 | 0.02 |
ENSDART00000181736
|
cant1b
|
calcium activated nucleotidase 1b |
| chr11_+_43431289 | 0.02 |
ENSDART00000192526
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr2_+_11685742 | 0.02 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr13_+_48358467 | 0.02 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr5_-_28109416 | 0.02 |
ENSDART00000042002
|
si:ch211-48m9.1
|
si:ch211-48m9.1 |
| chr17_-_10122204 | 0.02 |
ENSDART00000160751
|
BX088587.1
|
|
| chr5_-_38451082 | 0.02 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr24_+_9693951 | 0.02 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr14_-_4556896 | 0.02 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr19_+_19032141 | 0.02 |
ENSDART00000192432
|
cpne4b
|
copine IVb |
| chr19_-_1929310 | 0.02 |
ENSDART00000144113
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr6_+_11250316 | 0.01 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr3_-_13921173 | 0.01 |
ENSDART00000159177
ENSDART00000165174 |
si:dkey-61n16.5
|
si:dkey-61n16.5 |
| chr23_-_21453614 | 0.01 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr15_-_14552101 | 0.01 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr12_+_20641471 | 0.01 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr13_-_12602920 | 0.01 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr13_+_12299997 | 0.01 |
ENSDART00000108535
|
gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| chr10_-_5581487 | 0.01 |
ENSDART00000141943
|
syk
|
spleen tyrosine kinase |
| chr7_-_72261721 | 0.01 |
ENSDART00000172229
|
RASGRP2
|
RAS guanyl releasing protein 2 |
| chr19_-_42045372 | 0.01 |
ENSDART00000144275
|
trioa
|
trio Rho guanine nucleotide exchange factor a |
| chr18_+_41527877 | 0.01 |
ENSDART00000146972
|
selenot1b
|
selenoprotein T, 1b |
| chr2_-_17044959 | 0.01 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr10_+_21650828 | 0.00 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr2_-_37043905 | 0.00 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr22_+_1615691 | 0.00 |
ENSDART00000171555
|
si:ch211-255f4.13
|
si:ch211-255f4.13 |
| chr5_-_23280098 | 0.00 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr20_-_23842631 | 0.00 |
ENSDART00000153079
|
si:dkey-15j16.3
|
si:dkey-15j16.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0034397 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.6 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.6 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:2000660 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.7 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.1 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.5 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016841 | folic acid binding(GO:0005542) ammonia-lyase activity(GO:0016841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |