Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
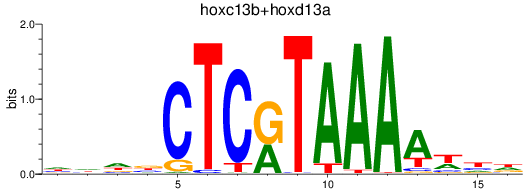
Results for hoxc13b+hoxd13a
Z-value: 0.68
Transcription factors associated with hoxc13b+hoxd13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxd13a
|
ENSDARG00000059256 | homeobox D13a |
|
hoxc13b
|
ENSDARG00000113877 | homeobox C13b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc13b | dr11_v1_chr11_+_2156430_2156430 | -0.85 | 9.9e-06 | Click! |
| hoxd13a | dr11_v1_chr9_-_1990323_1990323 | -0.04 | 8.7e-01 | Click! |
Activity profile of hoxc13b+hoxd13a motif
Sorted Z-values of hoxc13b+hoxd13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_27550768 | 2.13 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr6_-_8360918 | 2.10 |
ENSDART00000004716
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr7_-_48263516 | 1.97 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr22_-_22164338 | 1.92 |
ENSDART00000183840
|
cdc34a
|
cell division cycle 34 homolog (S. cerevisiae) a |
| chr17_-_4245311 | 1.78 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr17_-_4245902 | 1.52 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr3_+_43102010 | 1.35 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr9_+_1365747 | 1.34 |
ENSDART00000140917
ENSDART00000036605 |
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr1_+_24387659 | 1.33 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr23_+_24501918 | 1.22 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr4_-_4535189 | 1.19 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr3_+_23045214 | 1.17 |
ENSDART00000157090
ENSDART00000156472 |
b4galnt2.2
|
beta-1,4-N-acetyl-galactosaminyl transferase 2, tandem duplicate 2 |
| chr24_+_5799764 | 1.16 |
ENSDART00000154482
|
si:ch211-157j23.2
|
si:ch211-157j23.2 |
| chr2_+_37295088 | 1.13 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr14_+_8940326 | 1.12 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr16_+_39146696 | 1.12 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr5_+_57480014 | 1.09 |
ENSDART00000135520
|
si:ch211-202f5.3
|
si:ch211-202f5.3 |
| chr19_+_20201254 | 1.09 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr17_+_28005763 | 1.05 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr19_+_20201593 | 1.04 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr4_-_77551860 | 1.02 |
ENSDART00000188176
|
AL935186.6
|
|
| chr14_+_16083818 | 1.00 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr12_-_27212880 | 0.99 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr9_-_16239032 | 0.97 |
ENSDART00000131398
ENSDART00000138204 |
myo1b
|
myosin IB |
| chr8_-_410728 | 0.95 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr2_-_6065416 | 0.95 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr5_-_69621227 | 0.95 |
ENSDART00000178543
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr13_-_12389748 | 0.94 |
ENSDART00000141606
|
commd8
|
COMM domain containing 8 |
| chr20_+_14789305 | 0.93 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr15_+_23722620 | 0.93 |
ENSDART00000011447
|
sae1
|
SUMO1 activating enzyme subunit 1 |
| chr22_+_10676981 | 0.92 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr15_-_14194208 | 0.89 |
ENSDART00000188237
ENSDART00000183155 ENSDART00000165520 |
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr3_-_15475067 | 0.89 |
ENSDART00000025324
ENSDART00000139575 |
spns1
|
spinster homolog 1 (Drosophila) |
| chr19_-_23249822 | 0.89 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr24_+_17334682 | 0.84 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr16_+_32736588 | 0.84 |
ENSDART00000075191
ENSDART00000168358 |
zgc:172323
|
zgc:172323 |
| chr14_+_21699129 | 0.84 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr9_+_23003208 | 0.83 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr15_-_14193926 | 0.83 |
ENSDART00000162707
|
pnkp
|
polynucleotide kinase 3'-phosphatase |
| chr2_+_26240339 | 0.82 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr2_-_37353098 | 0.81 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr13_+_42309688 | 0.80 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr15_-_20412286 | 0.80 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr6_-_52661824 | 0.79 |
ENSDART00000149343
|
gdf5
|
growth differentiation factor 5 |
| chr23_+_9508538 | 0.79 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr25_-_25142387 | 0.77 |
ENSDART00000031814
|
tsg101a
|
tumor susceptibility 101a |
| chr16_+_33143503 | 0.76 |
ENSDART00000058471
ENSDART00000179385 |
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr3_+_46764278 | 0.75 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr15_-_6966221 | 0.75 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr25_-_20666754 | 0.75 |
ENSDART00000158418
|
csk
|
C-terminal Src kinase |
| chr5_-_24008997 | 0.74 |
ENSDART00000066645
|
eif1axa
|
eukaryotic translation initiation factor 1A, X-linked, a |
| chr19_+_24068223 | 0.74 |
ENSDART00000141351
ENSDART00000100420 |
pex11b
|
peroxisomal biogenesis factor 11 beta |
| chr12_+_17154655 | 0.73 |
ENSDART00000028003
|
ankrd22
|
ankyrin repeat domain 22 |
| chr5_-_32489796 | 0.73 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr2_+_26240631 | 0.72 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr17_-_41798856 | 0.71 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr16_-_12060770 | 0.71 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr9_-_29003245 | 0.70 |
ENSDART00000183391
ENSDART00000188836 |
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr5_-_69620722 | 0.69 |
ENSDART00000097248
|
aldh2.2
|
aldehyde dehydrogenase 2 family (mitochondrial), tandem duplicate 2 |
| chr3_-_40254634 | 0.69 |
ENSDART00000154562
|
top3a
|
DNA topoisomerase III alpha |
| chr7_+_24881680 | 0.69 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr11_+_31323746 | 0.69 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr15_+_23657051 | 0.69 |
ENSDART00000078336
|
klc3
|
kinesin light chain 3 |
| chr11_+_19271557 | 0.69 |
ENSDART00000190559
|
prickle2b
|
prickle homolog 2b |
| chr14_-_33978117 | 0.67 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr19_+_43669122 | 0.66 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr5_+_4006837 | 0.66 |
ENSDART00000138862
|
pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr14_+_35464994 | 0.65 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr24_-_25166416 | 0.65 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr17_+_13099476 | 0.64 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr6_+_13207139 | 0.64 |
ENSDART00000185601
ENSDART00000182182 |
ino80db
|
INO80 complex subunit Db |
| chr17_-_24938845 | 0.64 |
ENSDART00000156953
|
CR391986.1
|
|
| chr5_-_33280699 | 0.62 |
ENSDART00000183838
|
kyat1
|
kynurenine aminotransferase 1 |
| chr19_+_42227400 | 0.62 |
ENSDART00000131574
ENSDART00000135436 |
jtb
|
jumping translocation breakpoint |
| chr16_-_12060488 | 0.61 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr3_+_46763745 | 0.61 |
ENSDART00000185437
|
prkcsh
|
protein kinase C substrate 80K-H |
| chr14_+_26247319 | 0.61 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr18_-_25051846 | 0.60 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr17_+_8754020 | 0.60 |
ENSDART00000105322
|
edrf1
|
erythroid differentiation regulatory factor 1 |
| chr13_-_37474989 | 0.59 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr6_+_49771626 | 0.58 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr6_-_34838397 | 0.58 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr25_-_20666328 | 0.58 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr24_-_10393969 | 0.58 |
ENSDART00000106260
|
ankha
|
ANKH inorganic pyrophosphate transport regulator a |
| chr3_+_31058464 | 0.56 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr16_+_33144306 | 0.56 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr5_+_37504309 | 0.56 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr9_-_43207768 | 0.56 |
ENSDART00000192523
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr5_-_12031174 | 0.56 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr11_+_41838036 | 0.55 |
ENSDART00000192352
|
akr7a3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr19_-_31007417 | 0.55 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr25_-_20049449 | 0.55 |
ENSDART00000104315
|
zgc:136858
|
zgc:136858 |
| chr8_+_23861461 | 0.55 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr3_-_31086770 | 0.55 |
ENSDART00000103421
|
zgc:153292
|
zgc:153292 |
| chr1_-_493218 | 0.54 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr4_+_13901458 | 0.54 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr5_+_51111766 | 0.53 |
ENSDART00000188552
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr19_+_29808699 | 0.53 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr3_+_35005730 | 0.53 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr23_-_37291793 | 0.53 |
ENSDART00000083281
ENSDART00000187108 |
mul1b
|
mitochondrial E3 ubiquitin protein ligase 1b |
| chr20_-_42241150 | 0.52 |
ENSDART00000142791
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr17_+_12942634 | 0.52 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr8_-_54216499 | 0.52 |
ENSDART00000193678
|
mbd4
|
methyl-CpG binding domain protein 4 |
| chr5_+_27267186 | 0.51 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr23_+_33934228 | 0.51 |
ENSDART00000134237
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr3_+_35005062 | 0.50 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr2_-_37874647 | 0.50 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr13_+_35856463 | 0.50 |
ENSDART00000171056
ENSDART00000017202 |
kcnk1b
|
potassium channel, subfamily K, member 1b |
| chr18_+_8812549 | 0.50 |
ENSDART00000017619
|
impdh1a
|
IMP (inosine 5'-monophosphate) dehydrogenase 1a |
| chr8_-_25814263 | 0.49 |
ENSDART00000143397
|
taf10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr17_+_44441042 | 0.49 |
ENSDART00000142123
|
ap5m1
|
adaptor-related protein complex 5, mu 1 subunit |
| chr14_+_2487672 | 0.49 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr19_+_29808471 | 0.49 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr19_+_38167468 | 0.48 |
ENSDART00000160756
|
phf14
|
PHD finger protein 14 |
| chr7_+_13582256 | 0.48 |
ENSDART00000158477
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr14_+_16036139 | 0.47 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr19_+_7424347 | 0.46 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr11_-_26832685 | 0.46 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr2_-_11504778 | 0.46 |
ENSDART00000186556
|
sdr16c5a
|
short chain dehydrogenase/reductase family 16C, member 5a |
| chr21_+_37513058 | 0.46 |
ENSDART00000141096
|
amot
|
angiomotin |
| chr10_+_6907715 | 0.46 |
ENSDART00000041068
|
slc38a9
|
solute carrier family 38, member 9 |
| chr11_+_2649664 | 0.46 |
ENSDART00000166357
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr6_+_49771372 | 0.46 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr21_-_38730557 | 0.45 |
ENSDART00000150984
ENSDART00000111885 |
taf9
|
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr4_-_20108833 | 0.45 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr1_-_27014872 | 0.45 |
ENSDART00000147414
ENSDART00000134032 ENSDART00000192087 ENSDART00000189111 ENSDART00000187348 ENSDART00000187248 |
cntln
|
centlein, centrosomal protein |
| chr19_+_38168006 | 0.45 |
ENSDART00000087662
ENSDART00000177759 |
phf14
|
PHD finger protein 14 |
| chr16_-_27138478 | 0.45 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr19_+_40122160 | 0.44 |
ENSDART00000143966
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr21_+_37513488 | 0.44 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr5_+_30520249 | 0.44 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr1_+_38818268 | 0.43 |
ENSDART00000166864
|
spcs3
|
signal peptidase complex subunit 3 |
| chr3_-_20118342 | 0.43 |
ENSDART00000139902
|
selenow2a
|
selenoprotein W, 2a |
| chr1_+_45925150 | 0.43 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr15_+_32387063 | 0.43 |
ENSDART00000154210
ENSDART00000156525 |
si:ch211-162k9.5
|
si:ch211-162k9.5 |
| chr11_+_2649891 | 0.43 |
ENSDART00000093052
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr23_-_24343363 | 0.43 |
ENSDART00000166392
|
fam131c
|
family with sequence similarity 131, member C |
| chr5_-_67365006 | 0.43 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr4_-_13156971 | 0.42 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr2_-_37458527 | 0.42 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr17_-_44440832 | 0.42 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr10_+_37181780 | 0.42 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr3_-_25268751 | 0.42 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr2_+_24374305 | 0.42 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr23_-_4933508 | 0.42 |
ENSDART00000137578
ENSDART00000141196 |
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr1_+_45925365 | 0.42 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr1_-_59243542 | 0.42 |
ENSDART00000163021
|
mvb12a
|
multivesicular body subunit 12A |
| chr6_+_48206535 | 0.41 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr23_+_4226341 | 0.41 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr23_+_31596441 | 0.41 |
ENSDART00000053534
|
tbpl1
|
TBP-like 1 |
| chr7_-_19997095 | 0.41 |
ENSDART00000180955
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr13_-_34862452 | 0.41 |
ENSDART00000134573
ENSDART00000047552 |
sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr25_-_13871118 | 0.41 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr20_-_42241456 | 0.40 |
ENSDART00000034054
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr25_+_19913611 | 0.40 |
ENSDART00000155029
|
gramd4b
|
GRAM domain containing 4b |
| chr20_-_154989 | 0.39 |
ENSDART00000064542
|
rpf2
|
ribosome production factor 2 homolog |
| chr14_+_989733 | 0.39 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr22_-_19102256 | 0.39 |
ENSDART00000171866
ENSDART00000166295 |
polrmt
|
polymerase (RNA) mitochondrial (DNA directed) |
| chr17_+_30369107 | 0.39 |
ENSDART00000182286
ENSDART00000139091 |
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr10_+_24690534 | 0.38 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr23_+_19594608 | 0.38 |
ENSDART00000134865
|
slmapb
|
sarcolemma associated protein b |
| chr25_-_3867990 | 0.38 |
ENSDART00000075663
|
cracr2b
|
calcium release activated channel regulator 2B |
| chr21_+_30007566 | 0.38 |
ENSDART00000149124
ENSDART00000150071 |
ttc1
|
tetratricopeptide repeat domain 1 |
| chr5_-_67365333 | 0.37 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr9_-_12574473 | 0.37 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr5_-_67365750 | 0.37 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr12_-_17152139 | 0.37 |
ENSDART00000152478
|
stambpl1
|
STAM binding protein-like 1 |
| chr8_-_23758312 | 0.36 |
ENSDART00000132659
|
INAVA
|
si:ch211-163l21.4 |
| chr8_-_12432604 | 0.36 |
ENSDART00000133350
ENSDART00000140699 ENSDART00000101174 |
traf1
|
TNF receptor-associated factor 1 |
| chr6_-_39069244 | 0.36 |
ENSDART00000127365
|
eif4bb
|
eukaryotic translation initiation factor 4Bb |
| chr21_+_11749097 | 0.36 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr8_-_35960987 | 0.36 |
ENSDART00000160503
|
slc15a4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr20_+_26683933 | 0.36 |
ENSDART00000139852
ENSDART00000077751 |
foxq1b
|
forkhead box Q1b |
| chr16_+_7380463 | 0.36 |
ENSDART00000029727
ENSDART00000149086 |
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr16_+_33144112 | 0.35 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr10_+_35347993 | 0.35 |
ENSDART00000131350
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr19_-_25081711 | 0.35 |
ENSDART00000058513
|
xkr8.3
|
XK, Kell blood group complex subunit-related family, member 8, tandem duplicate 3 |
| chr7_+_5905091 | 0.35 |
ENSDART00000167099
|
CU459186.3
|
Histone H3.2 |
| chr23_-_12158685 | 0.34 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr10_+_25222367 | 0.34 |
ENSDART00000042767
|
grm5a
|
glutamate receptor, metabotropic 5a |
| chr22_-_24297510 | 0.34 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr3_-_48612078 | 0.34 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr22_-_9860792 | 0.34 |
ENSDART00000155908
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr1_-_681116 | 0.31 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr18_+_6126506 | 0.31 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr4_+_77966055 | 0.31 |
ENSDART00000174203
ENSDART00000130100 ENSDART00000080665 ENSDART00000174317 ENSDART00000190123 |
zgc:113921
|
zgc:113921 |
| chr20_+_4392687 | 0.30 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr7_+_13491452 | 0.30 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr3_+_45368973 | 0.29 |
ENSDART00000187282
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr22_+_38276024 | 0.29 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr3_+_13842554 | 0.29 |
ENSDART00000162317
ENSDART00000158068 |
ilf3b
|
interleukin enhancer binding factor 3b |
| chr18_-_35459605 | 0.28 |
ENSDART00000135691
|
itpkcb
|
inositol-trisphosphate 3-kinase Cb |
| chr19_+_20793237 | 0.28 |
ENSDART00000014774
|
txnl4a
|
thioredoxin-like 4A |
| chr5_+_61459422 | 0.28 |
ENSDART00000050902
|
polr2j
|
polymerase (RNA) II (DNA directed) polypeptide J |
| chr15_+_17722054 | 0.28 |
ENSDART00000191390
ENSDART00000169550 |
si:ch211-213d14.1
|
si:ch211-213d14.1 |
| chr7_+_39011852 | 0.27 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr23_-_4225830 | 0.27 |
ENSDART00000170455
|
aar2
|
AAR2 splicing factor homolog (S. cerevisiae) |
| chr4_-_26095755 | 0.27 |
ENSDART00000100611
ENSDART00000191266 |
si:ch211-244b2.3
|
si:ch211-244b2.3 |
| chr7_+_37742299 | 0.27 |
ENSDART00000143300
|
brd7
|
bromodomain containing 7 |
| chr22_+_35205968 | 0.27 |
ENSDART00000150467
|
tsc22d2
|
TSC22 domain family 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc13b+hoxd13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.4 | 2.0 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 0.9 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.3 | 1.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.3 | 1.3 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.2 | 1.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 1.1 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 1.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 2.0 | GO:0045453 | bone resorption(GO:0045453) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 0.8 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 1.0 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 0.4 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 1.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.9 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.4 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.4 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.9 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.3 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.9 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 0.5 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.4 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 1.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 2.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 1.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.2 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.4 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.8 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.4 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.9 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.2 | GO:0090266 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 1.1 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.1 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.6 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.4 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 0.2 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.5 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.5 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.8 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.4 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.8 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 2.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:2000622 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 1.0 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 1.6 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.1 | GO:1903430 | negative regulation of cell maturation(GO:1903430) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.9 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.7 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 1.0 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.5 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.3 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 1.9 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 1.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.8 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.6 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.0 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.2 | 1.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 0.9 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 0.5 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.6 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.8 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.7 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 0.9 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.4 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 1.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 1.2 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.4 | 1.2 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.3 | 1.3 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.3 | 0.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.3 | 1.2 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.3 | 2.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 0.9 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 0.6 | GO:0047804 | cysteine-S-conjugate beta-lyase activity(GO:0047804) |
| 0.2 | 0.6 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 1.0 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 1.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.8 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.6 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.2 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 1.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.5 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.4 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 1.0 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.9 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.4 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 0.5 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 0.4 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.3 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 2.0 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.2 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.5 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.0 | 3.1 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.1 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 2.0 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.8 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.6 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 3.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.1 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID INSULIN PATHWAY | Insulin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 1.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.8 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.1 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.5 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.0 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 2.4 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |