Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for hoxc13a
Z-value: 0.72
Transcription factors associated with hoxc13a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc13a
|
ENSDARG00000070353 | homeobox C13a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc13a | dr11_v1_chr23_+_36052944_36052944 | -0.79 | 8.3e-05 | Click! |
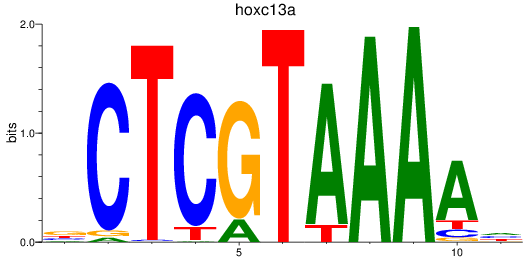
Activity profile of hoxc13a motif
Sorted Z-values of hoxc13a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_20201254 | 1.63 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr19_+_20201593 | 1.36 |
ENSDART00000163026
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr7_-_48263516 | 1.33 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr9_+_1365747 | 1.16 |
ENSDART00000140917
ENSDART00000036605 |
prkra
|
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr7_-_26518086 | 1.16 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr14_-_7045009 | 0.92 |
ENSDART00000112082
|
rufy1
|
RUN and FYVE domain containing 1 |
| chr20_-_2713120 | 0.86 |
ENSDART00000138753
ENSDART00000104606 |
rars2
|
arginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr3_+_46764278 | 0.82 |
ENSDART00000136051
ENSDART00000164930 |
prkcsh
|
protein kinase C substrate 80K-H |
| chr7_+_5905091 | 0.82 |
ENSDART00000167099
|
CU459186.3
|
Histone H3.2 |
| chr25_+_3294150 | 0.81 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr6_-_14038804 | 0.80 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr22_-_3299100 | 0.77 |
ENSDART00000160305
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr20_+_48116476 | 0.76 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr18_-_43866001 | 0.71 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr22_-_3299355 | 0.71 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr4_-_2727491 | 0.70 |
ENSDART00000141760
ENSDART00000039083 ENSDART00000134442 |
slco1c1
|
solute carrier organic anion transporter family, member 1C1 |
| chr18_-_43866526 | 0.70 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr13_+_8892784 | 0.69 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr18_+_14633974 | 0.66 |
ENSDART00000133834
|
vps9d1
|
VPS9 domain containing 1 |
| chr14_+_2487672 | 0.65 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr10_+_24690534 | 0.63 |
ENSDART00000079549
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr6_+_49771626 | 0.62 |
ENSDART00000134207
|
ctsz
|
cathepsin Z |
| chr17_-_8673567 | 0.56 |
ENSDART00000192714
ENSDART00000012546 |
ctbp2a
|
C-terminal binding protein 2a |
| chr7_+_24114694 | 0.55 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr5_-_12031174 | 0.54 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr9_+_45428041 | 0.53 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr17_-_8673278 | 0.51 |
ENSDART00000171850
ENSDART00000017337 ENSDART00000148504 ENSDART00000148808 |
ctbp2a
|
C-terminal binding protein 2a |
| chr2_+_24374305 | 0.50 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr17_+_31221761 | 0.49 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr20_-_154989 | 0.49 |
ENSDART00000064542
|
rpf2
|
ribosome production factor 2 homolog |
| chr10_+_42589391 | 0.49 |
ENSDART00000067689
ENSDART00000075259 |
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr6_+_49771372 | 0.48 |
ENSDART00000063251
|
ctsz
|
cathepsin Z |
| chr10_+_42589707 | 0.47 |
ENSDART00000075269
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr20_-_27190393 | 0.45 |
ENSDART00000149024
|
btbd7
|
BTB (POZ) domain containing 7 |
| chr22_+_24214665 | 0.43 |
ENSDART00000163980
ENSDART00000167996 |
glrx2
|
glutaredoxin 2 |
| chr12_-_27212880 | 0.43 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr19_+_34230108 | 0.42 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr23_-_12158685 | 0.42 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr21_+_42226113 | 0.41 |
ENSDART00000170362
|
GABRB2 (1 of many)
|
gamma-aminobutyric acid type A receptor beta2 subunit |
| chr7_+_29890292 | 0.41 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr14_+_16083818 | 0.41 |
ENSDART00000168462
|
rnf103
|
ring finger protein 103 |
| chr20_-_31252809 | 0.41 |
ENSDART00000137236
|
hpcal1
|
hippocalcin-like 1 |
| chr13_+_7241170 | 0.39 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr7_-_46777876 | 0.39 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr9_-_12574473 | 0.39 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr19_+_366034 | 0.38 |
ENSDART00000093383
|
vps72
|
vacuolar protein sorting 72 homolog (S. cerevisiae) |
| chr4_-_73488406 | 0.37 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr22_+_10676981 | 0.37 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr14_-_26425416 | 0.37 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr23_+_9508538 | 0.36 |
ENSDART00000010697
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr1_-_25144439 | 0.36 |
ENSDART00000132355
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr9_-_33608427 | 0.36 |
ENSDART00000100849
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr2_-_37353098 | 0.36 |
ENSDART00000056522
|
skila
|
SKI-like proto-oncogene a |
| chr6_+_13206516 | 0.35 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr17_+_13099476 | 0.35 |
ENSDART00000012670
|
pnn
|
pinin, desmosome associated protein |
| chr9_-_25443094 | 0.34 |
ENSDART00000105492
|
acvr2aa
|
activin A receptor type 2Aa |
| chr13_-_25774183 | 0.34 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr19_+_43017931 | 0.33 |
ENSDART00000132213
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr9_-_41088279 | 0.33 |
ENSDART00000000564
|
asnsd1
|
asparagine synthetase domain containing 1 |
| chr10_-_3416258 | 0.33 |
ENSDART00000005168
|
ddx55
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 |
| chr22_-_24297510 | 0.32 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr22_+_24215007 | 0.32 |
ENSDART00000162227
|
glrx2
|
glutaredoxin 2 |
| chr5_-_19014589 | 0.29 |
ENSDART00000002624
|
ranbp1
|
RAN binding protein 1 |
| chr23_+_33934228 | 0.28 |
ENSDART00000134237
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr20_+_33744065 | 0.28 |
ENSDART00000015000
|
henmt1
|
HEN methyltransferase 1 |
| chr14_+_35464994 | 0.27 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr7_-_6467510 | 0.27 |
ENSDART00000166041
|
FP325123.1
|
Histone H3.2 |
| chr25_-_13871118 | 0.27 |
ENSDART00000160866
|
cry2
|
cryptochrome circadian clock 2 |
| chr12_-_22400999 | 0.26 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr24_-_6158933 | 0.26 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr3_+_35005730 | 0.26 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr8_-_28449782 | 0.24 |
ENSDART00000062702
|
cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr3_+_35005062 | 0.24 |
ENSDART00000181163
|
prkcbb
|
protein kinase C, beta b |
| chr25_+_28776562 | 0.23 |
ENSDART00000109702
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr3_+_31058464 | 0.23 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr3_+_29640996 | 0.23 |
ENSDART00000011052
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr19_-_31007417 | 0.22 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr5_-_71838520 | 0.22 |
ENSDART00000174396
|
CU927890.1
|
|
| chr3_-_48612078 | 0.22 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr9_+_38967998 | 0.20 |
ENSDART00000135581
|
map2
|
microtubule-associated protein 2 |
| chr16_-_4203022 | 0.18 |
ENSDART00000018686
|
rrp15
|
ribosomal RNA processing 15 homolog |
| chr19_+_30884960 | 0.18 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr25_+_31276842 | 0.18 |
ENSDART00000187238
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr11_+_31730680 | 0.18 |
ENSDART00000145497
|
diaph3
|
diaphanous-related formin 3 |
| chr14_-_5546497 | 0.16 |
ENSDART00000054877
|
fgf24
|
fibroblast growth factor 24 |
| chr15_-_24883956 | 0.16 |
ENSDART00000113199
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr6_-_19664848 | 0.15 |
ENSDART00000159749
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr16_+_33144306 | 0.15 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr6_+_42338309 | 0.13 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr5_+_21891305 | 0.13 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr6_-_19665114 | 0.12 |
ENSDART00000168985
|
usp43a
|
ubiquitin specific peptidase 43a |
| chr9_+_22003942 | 0.11 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr9_+_21990095 | 0.11 |
ENSDART00000146829
ENSDART00000133515 ENSDART00000193582 |
si:dkey-57a22.13
|
si:dkey-57a22.13 |
| chr3_+_23691847 | 0.11 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr4_-_55227527 | 0.10 |
ENSDART00000184323
ENSDART00000148091 |
si:dkey-61p9.9
|
si:dkey-61p9.9 |
| chr21_-_21537452 | 0.09 |
ENSDART00000142549
|
or133-7
|
odorant receptor, family H, subfamily 133, member 7 |
| chr18_+_17151916 | 0.09 |
ENSDART00000175488
|
trhr2
|
thyrotropin releasing hormone receptor 2 |
| chr13_+_40729618 | 0.09 |
ENSDART00000074829
|
ankrd2
|
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr10_-_34867401 | 0.09 |
ENSDART00000145545
|
dclk1a
|
doublecortin-like kinase 1a |
| chr3_-_7656059 | 0.08 |
ENSDART00000170917
|
junbb
|
JunB proto-oncogene, AP-1 transcription factor subunit b |
| chr2_-_56655769 | 0.08 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr25_+_19947298 | 0.08 |
ENSDART00000067648
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr15_-_917274 | 0.08 |
ENSDART00000156624
|
si:dkey-77f5.15
|
si:dkey-77f5.15 |
| chr10_+_37927100 | 0.07 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr24_+_25032340 | 0.07 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr16_-_17541890 | 0.07 |
ENSDART00000131328
|
clcn1b
|
chloride channel, voltage-sensitive 1b |
| chr21_-_21594841 | 0.07 |
ENSDART00000132490
|
or133-2
|
odorant receptor, family H, subfamily 133, member 2 |
| chr25_-_31739309 | 0.07 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr12_-_3828854 | 0.07 |
ENSDART00000158529
|
taok2b
|
TAO kinase 2b |
| chr1_+_18965750 | 0.06 |
ENSDART00000132379
|
limch1a
|
LIM and calponin homology domains 1a |
| chr6_-_54816567 | 0.06 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr15_+_27387555 | 0.06 |
ENSDART00000018603
|
tbx4
|
T-box 4 |
| chr25_-_7887274 | 0.06 |
ENSDART00000157276
|
si:ch73-151m17.5
|
si:ch73-151m17.5 |
| chr16_-_26296477 | 0.05 |
ENSDART00000157553
|
erfl1
|
Ets2 repressor factor like 1 |
| chr1_-_17803614 | 0.05 |
ENSDART00000138475
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr3_+_59851537 | 0.05 |
ENSDART00000180997
|
CU693479.1
|
|
| chr24_+_1294176 | 0.05 |
ENSDART00000106637
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr9_+_25776194 | 0.04 |
ENSDART00000144499
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_+_33144112 | 0.04 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_+_36497250 | 0.04 |
ENSDART00000184727
|
TENM3
|
si:dkey-237h12.3 |
| chr25_+_31277415 | 0.04 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr6_+_49412754 | 0.04 |
ENSDART00000027398
|
kcna2a
|
potassium voltage-gated channel, shaker-related subfamily, member 2a |
| chr9_-_1959917 | 0.04 |
ENSDART00000082359
|
hoxd3a
|
homeobox D3a |
| chr4_+_37485430 | 0.03 |
ENSDART00000182667
|
si:dkeyp-86c4.1
|
si:dkeyp-86c4.1 |
| chr6_+_52869892 | 0.03 |
ENSDART00000146143
|
si:dkeyp-3f10.16
|
si:dkeyp-3f10.16 |
| chr25_-_3892686 | 0.03 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr8_+_14792830 | 0.03 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr4_+_41835660 | 0.02 |
ENSDART00000171428
|
si:ch211-268b14.2
|
si:ch211-268b14.2 |
| chr20_-_27086143 | 0.02 |
ENSDART00000008590
|
itpk1a
|
inositol-tetrakisphosphate 1-kinase a |
| chr14_+_23811808 | 0.02 |
ENSDART00000014411
|
kctd16a
|
potassium channel tetramerization domain containing 16a |
| chr15_-_24884784 | 0.01 |
ENSDART00000181808
|
aipl1
|
aryl hydrocarbon receptor interacting protein-like 1 |
| chr1_-_22370660 | 0.01 |
ENSDART00000127506
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr7_+_10563017 | 0.01 |
ENSDART00000193520
ENSDART00000173125 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr11_-_16093018 | 0.00 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr9_-_33749556 | 0.00 |
ENSDART00000149383
|
ndp
|
Norrie disease (pseudoglioma) |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc13a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.3 | 1.3 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.2 | 1.4 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 1.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.1 | 0.5 | GO:0007100 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.1 | 1.0 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 1.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 3.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.2 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.8 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.8 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 1.0 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.7 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.6 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.5 | GO:0061138 | morphogenesis of a branching epithelium(GO:0061138) |
| 0.0 | 0.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.4 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.6 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.2 | 0.5 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 0.8 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.4 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.3 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.2 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 3.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 1.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.4 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 1.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 0.7 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 3.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |