Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
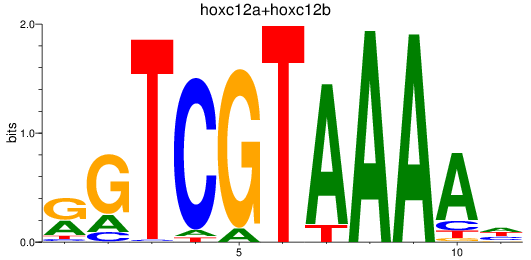
Results for hoxc12a+hoxc12b
Z-value: 0.42
Transcription factors associated with hoxc12a+hoxc12b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc12a
|
ENSDARG00000070352 | homeobox C12a |
|
hoxc12b
|
ENSDARG00000103133 | homeobox C12b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc12b | dr11_v1_chr11_+_2172335_2172335 | -0.27 | 2.7e-01 | Click! |
| hoxc12a | dr11_v1_chr23_+_36063599_36063599 | -0.09 | 7.3e-01 | Click! |
Activity profile of hoxc12a+hoxc12b motif
Sorted Z-values of hoxc12a+hoxc12b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_1550709 | 2.19 |
ENSDART00000110097
|
si:ch73-303b9.1
|
si:ch73-303b9.1 |
| chr8_+_52442785 | 1.32 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr2_-_17114852 | 1.22 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr7_+_46019780 | 1.00 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr18_+_18000887 | 0.86 |
ENSDART00000147797
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr10_-_31805923 | 0.85 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr5_-_32489796 | 0.78 |
ENSDART00000168870
|
gpr107
|
G protein-coupled receptor 107 |
| chr9_+_22634073 | 0.73 |
ENSDART00000181822
|
etv5a
|
ets variant 5a |
| chr6_-_13709591 | 0.71 |
ENSDART00000151771
|
chpfb
|
chondroitin polymerizing factor b |
| chr6_+_2174082 | 0.71 |
ENSDART00000073936
|
acvr1bb
|
activin A receptor type 1Bb |
| chr15_-_28904371 | 0.71 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr1_-_29061285 | 0.67 |
ENSDART00000053933
ENSDART00000142350 ENSDART00000192615 |
gemin8
|
gem (nuclear organelle) associated protein 8 |
| chr17_+_17764979 | 0.66 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr6_-_14038804 | 0.64 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr20_-_44576949 | 0.63 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr15_+_29024895 | 0.62 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr9_-_29039506 | 0.60 |
ENSDART00000100744
|
tmem177
|
transmembrane protein 177 |
| chr2_-_55797318 | 0.60 |
ENSDART00000158147
|
calr3b
|
calreticulin 3b |
| chr8_+_23861461 | 0.58 |
ENSDART00000037109
|
srpk1a
|
SRSF protein kinase 1a |
| chr13_+_33268657 | 0.58 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr2_-_6065416 | 0.57 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr13_+_8892784 | 0.57 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr21_-_22357985 | 0.57 |
ENSDART00000101751
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr3_+_16663373 | 0.56 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr11_-_6880725 | 0.51 |
ENSDART00000007204
|
ddx49
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 |
| chr19_+_26640096 | 0.50 |
ENSDART00000067793
|
ints3
|
integrator complex subunit 3 |
| chr1_-_26782573 | 0.50 |
ENSDART00000090611
|
sh3gl2a
|
SH3 domain containing GRB2 like 2a, endophilin A1 |
| chr1_-_38171648 | 0.50 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr15_-_20412286 | 0.49 |
ENSDART00000008589
|
chp2
|
calcineurin-like EF-hand protein 2 |
| chr13_-_25196758 | 0.48 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr22_-_31517300 | 0.46 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr5_-_24542726 | 0.46 |
ENSDART00000182975
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr5_+_41477954 | 0.46 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr5_-_54395488 | 0.46 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr17_+_49500820 | 0.45 |
ENSDART00000170306
|
AREL1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr3_-_27868183 | 0.45 |
ENSDART00000185812
|
abat
|
4-aminobutyrate aminotransferase |
| chr13_+_29925397 | 0.45 |
ENSDART00000123482
|
cuedc2
|
CUE domain containing 2 |
| chr25_+_3294150 | 0.45 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr3_-_40254634 | 0.43 |
ENSDART00000154562
|
top3a
|
DNA topoisomerase III alpha |
| chr12_-_17199381 | 0.43 |
ENSDART00000193292
|
lipf
|
lipase, gastric |
| chr7_-_25133783 | 0.43 |
ENSDART00000173781
ENSDART00000121943 ENSDART00000077219 |
badb
|
BCL2 associated agonist of cell death b |
| chr6_-_3992942 | 0.41 |
ENSDART00000182328
|
unc50
|
unc-50 homolog (C. elegans) |
| chr13_-_3936555 | 0.41 |
ENSDART00000017052
|
ncoa4
|
nuclear receptor coactivator 4 |
| chr3_-_23575007 | 0.40 |
ENSDART00000155282
ENSDART00000087726 |
igf2bp1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr6_-_34838397 | 0.40 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr11_+_43751263 | 0.40 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr7_-_30624435 | 0.39 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr6_+_27991943 | 0.39 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr5_-_9216758 | 0.39 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr22_+_10676981 | 0.38 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr3_-_18737126 | 0.38 |
ENSDART00000055767
|
e4f1
|
E4F transcription factor 1 |
| chr6_+_13207139 | 0.38 |
ENSDART00000185601
ENSDART00000182182 |
ino80db
|
INO80 complex subunit Db |
| chr18_-_21685055 | 0.36 |
ENSDART00000019861
|
mbtps1
|
membrane-bound transcription factor peptidase, site 1 |
| chr20_+_34671386 | 0.36 |
ENSDART00000152836
ENSDART00000138226 |
elp3
|
elongator acetyltransferase complex subunit 3 |
| chr12_-_11560794 | 0.35 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr2_+_31330358 | 0.35 |
ENSDART00000178066
|
clul1
|
clusterin-like 1 (retinal) |
| chr2_+_8261109 | 0.35 |
ENSDART00000024677
|
dia1a
|
deleted in autism 1a |
| chr6_-_53334259 | 0.33 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr10_+_15967643 | 0.33 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr25_-_37121335 | 0.33 |
ENSDART00000017805
|
nfat5a
|
nuclear factor of activated T cells 5a |
| chr13_+_38817871 | 0.32 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr19_+_7424347 | 0.32 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr13_-_36050303 | 0.31 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr2_+_23039041 | 0.31 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr18_-_31105391 | 0.31 |
ENSDART00000039495
|
pdcd5
|
programmed cell death 5 |
| chr6_+_27992886 | 0.31 |
ENSDART00000160354
|
amotl2a
|
angiomotin like 2a |
| chr11_-_26832685 | 0.30 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr3_-_48612078 | 0.30 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr2_+_55199721 | 0.29 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr19_+_30884960 | 0.29 |
ENSDART00000140603
ENSDART00000183224 ENSDART00000135484 ENSDART00000139599 |
yars
|
tyrosyl-tRNA synthetase |
| chr16_+_10429770 | 0.29 |
ENSDART00000173132
|
vars
|
valyl-tRNA synthetase |
| chr4_-_13156971 | 0.28 |
ENSDART00000182164
|
grip1
|
glutamate receptor interacting protein 1 |
| chr7_+_29167744 | 0.28 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr2_-_20715094 | 0.28 |
ENSDART00000155439
|
dusp12
|
dual specificity phosphatase 12 |
| chr2_-_37134169 | 0.27 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr5_-_31857345 | 0.27 |
ENSDART00000112546
|
pkn3
|
protein kinase N3 |
| chr9_+_38314466 | 0.27 |
ENSDART00000048753
|
ddx18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr15_-_28262632 | 0.26 |
ENSDART00000134601
ENSDART00000175022 |
prpf8
|
pre-mRNA processing factor 8 |
| chr24_-_24796583 | 0.26 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr6_+_3730843 | 0.25 |
ENSDART00000019630
|
FO704755.1
|
|
| chr6_+_42338309 | 0.25 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr24_-_17029374 | 0.25 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr3_+_31058464 | 0.24 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr4_-_73488406 | 0.24 |
ENSDART00000115002
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr5_+_32490238 | 0.23 |
ENSDART00000191839
|
ndor1
|
NADPH dependent diflavin oxidoreductase 1 |
| chr15_+_44093286 | 0.23 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr15_-_23942861 | 0.23 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr8_-_8346023 | 0.23 |
ENSDART00000164731
ENSDART00000188259 |
tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr25_-_7670616 | 0.23 |
ENSDART00000131583
ENSDART00000142794 |
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr5_-_31856681 | 0.22 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr1_+_31019165 | 0.22 |
ENSDART00000102210
|
itga6b
|
integrin, alpha 6b |
| chr22_-_10440688 | 0.22 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr7_+_30240791 | 0.21 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr16_-_12060488 | 0.21 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr2_-_37874647 | 0.20 |
ENSDART00000039386
|
zgc:66427
|
zgc:66427 |
| chr19_-_7272921 | 0.19 |
ENSDART00000102075
ENSDART00000132887 ENSDART00000130234 ENSDART00000193535 ENSDART00000136528 |
rxrba
|
retinoid x receptor, beta a |
| chr14_+_2487672 | 0.19 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr15_+_5132439 | 0.19 |
ENSDART00000010350
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr7_+_10563017 | 0.19 |
ENSDART00000193520
ENSDART00000173125 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr7_+_24522308 | 0.18 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr2_-_18830722 | 0.18 |
ENSDART00000165330
ENSDART00000165698 |
pbx1a
|
pre-B-cell leukemia homeobox 1a |
| chr25_-_7670391 | 0.17 |
ENSDART00000044970
|
bet1l
|
Bet1 golgi vesicular membrane trafficking protein-like |
| chr6_-_28222592 | 0.17 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr15_-_19443997 | 0.16 |
ENSDART00000114936
|
esamb
|
endothelial cell adhesion molecule b |
| chr22_+_17286841 | 0.16 |
ENSDART00000133176
|
FAM163A
|
si:dkey-171o17.8 |
| chr15_+_43799258 | 0.15 |
ENSDART00000122238
|
tyr
|
tyrosinase |
| chr8_-_65189 | 0.15 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr20_-_19530751 | 0.14 |
ENSDART00000148574
|
eif2b4
|
eukaryotic translation initiation factor 2B, subunit 4 delta |
| chr16_+_38659475 | 0.13 |
ENSDART00000023238
|
emc2
|
ER membrane protein complex subunit 2 |
| chr5_-_26764298 | 0.13 |
ENSDART00000189373
|
rnf181
|
ring finger protein 181 |
| chr18_-_21748808 | 0.13 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr2_+_54696042 | 0.12 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr16_+_11242443 | 0.12 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr24_-_24797455 | 0.11 |
ENSDART00000138741
|
pde7a
|
phosphodiesterase 7A |
| chr16_-_6205790 | 0.10 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr3_+_37787229 | 0.10 |
ENSDART00000075028
|
rps11
|
ribosomal protein S11 |
| chr20_-_47422469 | 0.10 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr9_-_10145795 | 0.10 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr14_-_26425416 | 0.09 |
ENSDART00000088690
|
lman2
|
lectin, mannose-binding 2 |
| chr25_-_7887274 | 0.09 |
ENSDART00000157276
|
si:ch73-151m17.5
|
si:ch73-151m17.5 |
| chr18_+_3140682 | 0.09 |
ENSDART00000166382
|
clns1a
|
chloride channel, nucleotide-sensitive, 1A |
| chr7_+_27898091 | 0.09 |
ENSDART00000186451
ENSDART00000173510 |
tub
|
tubby bipartite transcription factor |
| chr22_+_30137374 | 0.08 |
ENSDART00000187808
|
add3a
|
adducin 3 (gamma) a |
| chr13_+_45431660 | 0.08 |
ENSDART00000099950
|
syf2
|
SYF2 pre-mRNA-splicing factor |
| chr15_+_34675660 | 0.08 |
ENSDART00000009127
|
lta
|
lymphotoxin alpha (TNF superfamily, member 1) |
| chr9_-_38399432 | 0.08 |
ENSDART00000148268
|
znf142
|
zinc finger protein 142 |
| chr5_-_21065094 | 0.07 |
ENSDART00000143785
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr17_-_23727978 | 0.07 |
ENSDART00000079600
|
minpp1a
|
multiple inositol-polyphosphate phosphatase 1a |
| chr4_-_73520581 | 0.07 |
ENSDART00000171513
|
si:ch73-266f23.1
|
si:ch73-266f23.1 |
| chr2_+_30103285 | 0.07 |
ENSDART00000019417
|
dnajb6a
|
DnaJ (Hsp40) homolog, subfamily B, member 6a |
| chr9_-_9242777 | 0.06 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr24_-_6898302 | 0.06 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr12_-_30777540 | 0.06 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr24_+_34069675 | 0.06 |
ENSDART00000143995
|
si:ch211-190p8.2
|
si:ch211-190p8.2 |
| chr9_+_20483846 | 0.05 |
ENSDART00000192067
ENSDART00000145111 |
parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr3_+_23691847 | 0.05 |
ENSDART00000078453
|
hoxb7a
|
homeobox B7a |
| chr3_-_10749691 | 0.05 |
ENSDART00000183088
|
CU210890.1
|
|
| chr16_-_47426482 | 0.05 |
ENSDART00000148631
ENSDART00000149723 |
sept7b
|
septin 7b |
| chr6_-_54816567 | 0.05 |
ENSDART00000150079
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr14_+_32022272 | 0.05 |
ENSDART00000105760
|
zic6
|
zic family member 6 |
| chr18_+_31280984 | 0.04 |
ENSDART00000170285
ENSDART00000150608 ENSDART00000159720 |
def8
|
differentially expressed in FDCP 8 homolog (mouse) |
| chr16_-_33806390 | 0.04 |
ENSDART00000160671
|
rspo1
|
R-spondin 1 |
| chr20_-_8094718 | 0.04 |
ENSDART00000122902
|
si:ch211-232i5.3
|
si:ch211-232i5.3 |
| chr20_+_46183505 | 0.04 |
ENSDART00000060799
|
taar13b
|
trace amine associated receptor 13b |
| chr11_+_7214353 | 0.04 |
ENSDART00000156764
|
nwd1
|
NACHT and WD repeat domain containing 1 |
| chr24_-_21913426 | 0.04 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr12_-_9123352 | 0.04 |
ENSDART00000149655
|
adam8b
|
ADAM metallopeptidase domain 8b |
| chr7_-_32833153 | 0.04 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr23_-_4855122 | 0.03 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr23_-_39849155 | 0.03 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr1_-_8101495 | 0.03 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr13_-_3351708 | 0.03 |
ENSDART00000042875
|
si:ch73-193i2.2
|
si:ch73-193i2.2 |
| chr4_-_46915962 | 0.03 |
ENSDART00000169555
|
si:ch211-134c10.1
|
si:ch211-134c10.1 |
| chr3_-_15734358 | 0.03 |
ENSDART00000137325
|
mvp
|
major vault protein |
| chr16_+_42471455 | 0.03 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr22_-_9736050 | 0.02 |
ENSDART00000152919
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr7_-_2116512 | 0.02 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
| chr19_-_10425140 | 0.02 |
ENSDART00000145319
|
si:ch211-171h4.3
|
si:ch211-171h4.3 |
| chr5_+_36611128 | 0.02 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr9_+_54290896 | 0.02 |
ENSDART00000149175
|
pou4f3
|
POU class 4 homeobox 3 |
| chr23_-_12014931 | 0.02 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr1_+_58424507 | 0.02 |
ENSDART00000114197
|
si:ch73-236c18.3
|
si:ch73-236c18.3 |
| chr23_+_33752275 | 0.02 |
ENSDART00000007260
|
si:ch211-210c8.6
|
si:ch211-210c8.6 |
| chr14_-_32866351 | 0.01 |
ENSDART00000166133
ENSDART00000172545 |
ube2a
|
ubiquitin-conjugating enzyme E2A (RAD6 homolog) |
| chr1_+_36437585 | 0.01 |
ENSDART00000189182
|
pou4f2
|
POU class 4 homeobox 2 |
| chr13_+_39182099 | 0.01 |
ENSDART00000131434
|
fam135a
|
family with sequence similarity 135, member A |
| chr11_-_18557929 | 0.01 |
ENSDART00000110882
ENSDART00000181381 ENSDART00000189312 |
dido1
|
death inducer-obliterator 1 |
| chr4_-_38259998 | 0.01 |
ENSDART00000172389
|
znf1093
|
zinc finger protein 1093 |
| chr18_+_33264609 | 0.01 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr21_-_4539899 | 0.01 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr22_+_10440991 | 0.01 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr22_+_15308526 | 0.01 |
ENSDART00000188280
|
CU104797.1
|
|
| chr1_-_34612613 | 0.00 |
ENSDART00000004361
|
klf5a
|
Kruppel-like factor 5a |
| chr10_-_15910974 | 0.00 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr17_-_51199219 | 0.00 |
ENSDART00000154403
|
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr12_+_46696867 | 0.00 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc12a+hoxc12b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.1 | 0.4 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.1 | 1.2 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.4 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.5 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.1 | 0.7 | GO:0035909 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.5 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.6 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.4 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.3 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0051299 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.0 | 0.4 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.6 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.1 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.0 | 0.3 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.0 | 0.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.3 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 1.0 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 1.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.4 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 0.4 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.0 | GO:0051044 | regulation of cellular extravasation(GO:0002691) positive regulation of cellular extravasation(GO:0002693) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 1.2 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.0 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.7 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.1 | 0.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.5 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.3 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.1 | 0.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.6 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.2 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 0.8 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.0 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.5 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.4 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |