Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
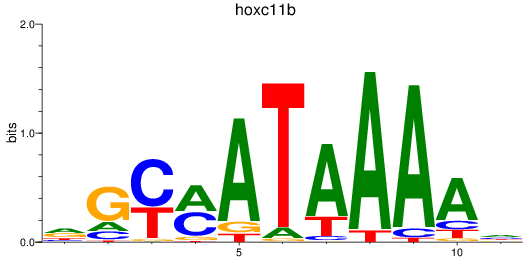
Results for hoxc11b
Z-value: 0.47
Transcription factors associated with hoxc11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxc11b
|
ENSDARG00000102631 | homeobox C11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc11b | dr11_v1_chr11_+_2180072_2180072 | -0.75 | 3.0e-04 | Click! |
Activity profile of hoxc11b motif
Sorted Z-values of hoxc11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_17029374 | 0.93 |
ENSDART00000039267
|
ptgdsb.1
|
prostaglandin D2 synthase b, tandem duplicate 1 |
| chr2_-_32738535 | 0.64 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr13_-_4018888 | 0.62 |
ENSDART00000058238
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr3_-_3366590 | 0.60 |
ENSDART00000109428
ENSDART00000175329 |
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr10_-_10969444 | 0.51 |
ENSDART00000138041
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr2_+_16597011 | 0.49 |
ENSDART00000160641
ENSDART00000125413 |
xrn1
|
5'-3' exoribonuclease 1 |
| chr14_+_4276394 | 0.48 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr10_-_10969596 | 0.47 |
ENSDART00000092011
|
exd3
|
exonuclease 3'-5' domain containing 3 |
| chr23_+_2666944 | 0.46 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr6_-_41138854 | 0.44 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr20_-_34670236 | 0.42 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr20_-_29499363 | 0.41 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr25_+_7532811 | 0.41 |
ENSDART00000161593
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr21_-_3700334 | 0.40 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr24_+_21720304 | 0.40 |
ENSDART00000147250
ENSDART00000048273 |
pan3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr16_-_42965192 | 0.39 |
ENSDART00000113714
|
mtx1a
|
metaxin 1a |
| chr9_+_21977383 | 0.38 |
ENSDART00000135032
|
si:dkey-57a22.11
|
si:dkey-57a22.11 |
| chr25_+_28823952 | 0.37 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr3_+_39579393 | 0.37 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr17_-_8592824 | 0.36 |
ENSDART00000127022
|
CU462878.1
|
|
| chr20_-_51831657 | 0.35 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr20_-_14462995 | 0.35 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr5_-_65662996 | 0.35 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr11_+_21586335 | 0.35 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr14_+_14841685 | 0.34 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr19_-_47571797 | 0.33 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr13_+_36585399 | 0.33 |
ENSDART00000030211
|
gmfb
|
glia maturation factor, beta |
| chr15_-_1843831 | 0.33 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr8_+_12951155 | 0.33 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr19_-_617246 | 0.33 |
ENSDART00000062551
|
cyp51
|
cytochrome P450, family 51 |
| chr13_-_25745089 | 0.32 |
ENSDART00000189333
ENSDART00000147420 |
sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr6_+_27667359 | 0.32 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr20_-_51831816 | 0.32 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr19_-_47571456 | 0.31 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr2_-_9971609 | 0.31 |
ENSDART00000137924
ENSDART00000048655 ENSDART00000131613 |
zgc:55943
|
zgc:55943 |
| chr20_+_27087539 | 0.30 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr15_-_16946124 | 0.30 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr15_+_44093286 | 0.30 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr24_-_32025637 | 0.30 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr14_-_36763302 | 0.30 |
ENSDART00000074786
|
ctso
|
cathepsin O |
| chr2_-_45663945 | 0.30 |
ENSDART00000075080
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr22_+_1953096 | 0.29 |
ENSDART00000166630
ENSDART00000186234 |
si:dkey-15h8.16
|
si:dkey-15h8.16 |
| chr12_-_44759246 | 0.28 |
ENSDART00000162888
|
dock1
|
dedicator of cytokinesis 1 |
| chr3_-_26183699 | 0.28 |
ENSDART00000147517
ENSDART00000140731 |
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr12_-_13337033 | 0.28 |
ENSDART00000105903
ENSDART00000139786 |
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr2_+_31948352 | 0.27 |
ENSDART00000192611
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr17_+_14965570 | 0.27 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr21_+_25236297 | 0.27 |
ENSDART00000112783
|
tmem45b
|
transmembrane protein 45B |
| chr11_+_37049105 | 0.27 |
ENSDART00000155408
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr11_+_37049347 | 0.26 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr3_+_35406998 | 0.25 |
ENSDART00000102994
|
rbbp6
|
retinoblastoma binding protein 6 |
| chr1_+_52481332 | 0.25 |
ENSDART00000074231
|
cldnd1b
|
claudin domain containing 1b |
| chr23_+_22785375 | 0.24 |
ENSDART00000142085
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr2_-_32366287 | 0.24 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr16_+_7380463 | 0.24 |
ENSDART00000029727
ENSDART00000149086 |
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr14_+_32918172 | 0.24 |
ENSDART00000182867
|
lnx2b
|
ligand of numb-protein X 2b |
| chr13_-_37465524 | 0.24 |
ENSDART00000100324
ENSDART00000147336 |
ppp2r5eb
|
protein phosphatase 2, regulatory subunit B', epsilon isoform b |
| chr16_+_13883872 | 0.24 |
ENSDART00000101304
ENSDART00000136005 |
atg12
|
ATG12 autophagy related 12 homolog (S. cerevisiae) |
| chr7_-_60096318 | 0.23 |
ENSDART00000189125
|
BX511067.1
|
|
| chr10_+_5234327 | 0.23 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr7_+_9981757 | 0.23 |
ENSDART00000113429
ENSDART00000173233 |
adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chr15_-_36660789 | 0.22 |
ENSDART00000150109
|
nphs1
|
nephrosis 1, congenital, Finnish type (nephrin) |
| chr3_+_52545014 | 0.22 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr3_+_52545400 | 0.21 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr8_+_23381892 | 0.21 |
ENSDART00000180950
ENSDART00000063010 ENSDART00000074241 ENSDART00000142783 |
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr16_-_21903083 | 0.21 |
ENSDART00000165849
|
setdb1b
|
SET domain, bifurcated 1b |
| chr17_+_15788100 | 0.21 |
ENSDART00000027667
|
rragd
|
ras-related GTP binding D |
| chr7_-_7692992 | 0.21 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr7_-_57637779 | 0.21 |
ENSDART00000028017
|
mad2l1
|
MAD2 mitotic arrest deficient-like 1 (yeast) |
| chr6_-_9282080 | 0.21 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr13_-_26799244 | 0.21 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr4_+_12292274 | 0.21 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr22_-_31517300 | 0.21 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr11_+_25112269 | 0.20 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr12_+_34770531 | 0.20 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr16_+_54829293 | 0.20 |
ENSDART00000024729
|
pabpc1a
|
poly(A) binding protein, cytoplasmic 1a |
| chr2_-_44777592 | 0.20 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr15_-_23936276 | 0.20 |
ENSDART00000137569
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr7_-_26076970 | 0.20 |
ENSDART00000101120
|
zgc:92664
|
zgc:92664 |
| chr25_+_14694876 | 0.20 |
ENSDART00000050478
ENSDART00000188093 |
mpped2
|
metallophosphoesterase domain containing 2b |
| chr11_-_12744082 | 0.19 |
ENSDART00000081319
|
zrsr2
|
zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2 |
| chr11_-_25257045 | 0.19 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr6_+_4528631 | 0.19 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr7_-_30280934 | 0.19 |
ENSDART00000126741
|
serf2
|
small EDRK-rich factor 2 |
| chr16_+_38940758 | 0.19 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr12_-_11560794 | 0.19 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr23_-_36003282 | 0.19 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr14_-_25949951 | 0.19 |
ENSDART00000141304
|
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr7_-_7692723 | 0.18 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr10_+_506538 | 0.18 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr5_-_21888368 | 0.18 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr5_+_44805028 | 0.18 |
ENSDART00000141198
|
ctsla
|
cathepsin La |
| chr10_+_32050906 | 0.18 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr20_-_48701593 | 0.17 |
ENSDART00000132835
|
pax1b
|
paired box 1b |
| chr7_+_10911396 | 0.17 |
ENSDART00000167273
ENSDART00000081323 ENSDART00000170655 |
abhd17c
|
abhydrolase domain containing 17C |
| chr8_+_36582728 | 0.17 |
ENSDART00000049230
|
pqbp1
|
polyglutamine binding protein 1 |
| chr6_+_27991943 | 0.17 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr1_-_45577980 | 0.17 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr16_-_6205790 | 0.16 |
ENSDART00000038495
|
ctnnb1
|
catenin (cadherin-associated protein), beta 1 |
| chr14_-_32959851 | 0.16 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr2_+_22409249 | 0.16 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr22_-_11648322 | 0.16 |
ENSDART00000109596
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr11_-_6265574 | 0.15 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr8_-_25033681 | 0.15 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr8_+_23382568 | 0.15 |
ENSDART00000129167
|
mapre1a
|
microtubule-associated protein, RP/EB family, member 1a |
| chr24_-_21511737 | 0.15 |
ENSDART00000109587
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr22_-_11648094 | 0.15 |
ENSDART00000191791
|
dpp4
|
dipeptidyl-peptidase 4 |
| chr21_-_39024754 | 0.15 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr20_-_25643667 | 0.15 |
ENSDART00000137457
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr21_-_19918286 | 0.15 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr6_+_58622831 | 0.15 |
ENSDART00000128793
|
sp7
|
Sp7 transcription factor |
| chr4_-_5652030 | 0.15 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr17_-_2690083 | 0.14 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr12_-_25380028 | 0.14 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr5_-_65037525 | 0.14 |
ENSDART00000158856
|
anxa1b
|
annexin A1b |
| chr11_-_25257595 | 0.14 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr7_+_46003449 | 0.14 |
ENSDART00000159700
ENSDART00000173625 |
si:ch211-260e23.9
|
si:ch211-260e23.9 |
| chr8_+_11687254 | 0.14 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr4_+_23125689 | 0.13 |
ENSDART00000077854
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr20_-_34028967 | 0.13 |
ENSDART00000153408
ENSDART00000033817 |
scyl3
|
SCY1-like, kinase-like 3 |
| chr8_+_47633438 | 0.13 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr3_-_9722603 | 0.13 |
ENSDART00000168234
|
crebbpb
|
CREB binding protein b |
| chr1_+_26105141 | 0.13 |
ENSDART00000102379
ENSDART00000127154 |
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr19_-_7033636 | 0.13 |
ENSDART00000122815
ENSDART00000123443 ENSDART00000124094 |
daxx
|
death-domain associated protein |
| chr1_+_47165842 | 0.13 |
ENSDART00000053152
ENSDART00000167051 |
cbr1
|
carbonyl reductase 1 |
| chr14_+_23184517 | 0.13 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr7_+_13639473 | 0.12 |
ENSDART00000157870
ENSDART00000032899 |
clpxa
|
caseinolytic mitochondrial matrix peptidase chaperone subunit a |
| chr12_-_20373058 | 0.12 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr24_+_5840258 | 0.12 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr11_+_24313931 | 0.12 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr10_-_36618674 | 0.12 |
ENSDART00000135302
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr3_+_62161184 | 0.12 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr13_-_25484659 | 0.12 |
ENSDART00000135321
ENSDART00000022799 |
tial1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr8_-_10961991 | 0.12 |
ENSDART00000139603
|
trim33
|
tripartite motif containing 33 |
| chr2_-_43196595 | 0.12 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr6_+_13083146 | 0.12 |
ENSDART00000172158
|
l3hypdh
|
trans-L-3-hydroxyproline dehydratase |
| chr19_+_9455218 | 0.11 |
ENSDART00000139385
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr12_+_10134366 | 0.11 |
ENSDART00000127343
|
smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr8_+_44677569 | 0.11 |
ENSDART00000191104
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr21_-_11970199 | 0.11 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr1_-_44933094 | 0.11 |
ENSDART00000147527
|
si:dkey-9i23.14
|
si:dkey-9i23.14 |
| chr25_+_30238021 | 0.11 |
ENSDART00000123220
|
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr9_+_2452672 | 0.10 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr18_-_15373620 | 0.10 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr2_+_21128391 | 0.10 |
ENSDART00000136814
|
pip4k2ab
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha b |
| chr5_-_57289872 | 0.10 |
ENSDART00000189893
ENSDART00000050957 |
fer
|
fer (fps/fes related) tyrosine kinase |
| chr15_-_25527580 | 0.10 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr10_-_25591194 | 0.10 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr1_+_54683655 | 0.10 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr6_+_35362225 | 0.10 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr12_+_19356623 | 0.10 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr7_-_44604821 | 0.10 |
ENSDART00000148967
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr6_-_31348999 | 0.10 |
ENSDART00000153734
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr8_-_45867358 | 0.10 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr24_-_29586082 | 0.09 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr19_-_35733401 | 0.09 |
ENSDART00000066712
|
trappc3
|
trafficking protein particle complex 3 |
| chr6_+_54221654 | 0.09 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr8_-_44298964 | 0.09 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr12_+_27704015 | 0.09 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr7_-_55051692 | 0.09 |
ENSDART00000170637
|
tpcn2
|
two pore segment channel 2 |
| chr18_+_619619 | 0.09 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr3_+_24134418 | 0.09 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr17_+_17764979 | 0.09 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr2_-_20788698 | 0.09 |
ENSDART00000181823
|
CABZ01020455.1
|
|
| chr16_+_10264601 | 0.08 |
ENSDART00000186167
|
mrs2
|
MRS2 magnesium transporter |
| chr3_+_54761569 | 0.08 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr3_-_19200571 | 0.08 |
ENSDART00000131503
ENSDART00000012335 |
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr11_-_38899816 | 0.08 |
ENSDART00000141515
|
pimr127
|
Pim proto-oncogene, serine/threonine kinase, related 127 |
| chr10_-_25328814 | 0.08 |
ENSDART00000123820
|
tmem135
|
transmembrane protein 135 |
| chr10_+_22775253 | 0.08 |
ENSDART00000190141
|
tmem88a
|
transmembrane protein 88 a |
| chr7_-_41420226 | 0.07 |
ENSDART00000074155
|
abcf2b
|
ATP-binding cassette, sub-family F (GCN20), member 2b |
| chr18_+_14342326 | 0.07 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr7_-_23745984 | 0.07 |
ENSDART00000048050
|
ITGB1BP2
|
zgc:92429 |
| chr8_-_14121634 | 0.07 |
ENSDART00000184946
|
bgna
|
biglycan a |
| chr18_-_41160975 | 0.07 |
ENSDART00000187517
|
CABZ01005876.1
|
|
| chr8_+_7737062 | 0.07 |
ENSDART00000166712
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr11_-_45152702 | 0.07 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr18_-_15559817 | 0.07 |
ENSDART00000061681
|
si:ch211-245j22.3
|
si:ch211-245j22.3 |
| chr1_+_2321384 | 0.07 |
ENSDART00000157662
|
farp1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr5_-_28767573 | 0.07 |
ENSDART00000158299
ENSDART00000043466 |
traf2a
|
Tnf receptor-associated factor 2a |
| chr25_+_20089986 | 0.07 |
ENSDART00000143441
ENSDART00000184073 |
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr21_+_21279159 | 0.07 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr2_+_23039041 | 0.07 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr18_-_17513426 | 0.07 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr12_-_33817114 | 0.06 |
ENSDART00000161265
|
twnk
|
twinkle mtDNA helicase |
| chr6_-_17206028 | 0.06 |
ENSDART00000155756
|
pimr3
|
Pim proto-oncogene, serine/threonine kinase, related 3 |
| chr2_+_19578446 | 0.06 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr2_+_19578079 | 0.06 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr2_+_19522082 | 0.06 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr21_+_22828500 | 0.06 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr5_+_45140914 | 0.06 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr24_+_40529346 | 0.05 |
ENSDART00000168548
|
CU929259.1
|
|
| chr4_+_27130412 | 0.05 |
ENSDART00000145083
|
brd1a
|
bromodomain containing 1a |
| chr21_+_10756154 | 0.05 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr22_+_2632961 | 0.05 |
ENSDART00000106278
|
znf1173
|
zinc finger protein 1173 |
| chr2_+_14992879 | 0.05 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr13_+_33268657 | 0.05 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr14_+_30279391 | 0.05 |
ENSDART00000172794
|
fgl1
|
fibrinogen-like 1 |
| chr23_+_36074798 | 0.05 |
ENSDART00000133760
ENSDART00000103146 |
hoxc11a
|
homeobox C11a |
| chr13_+_15933168 | 0.05 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxc11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 1.1 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.1 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0031652 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 0.4 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.2 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.2 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 0.2 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.3 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.2 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.7 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.6 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.5 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.4 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.1 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0031392 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0009791 | post-embryonic development(GO:0009791) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0071174 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.1 | GO:0034354 | tryptophan catabolic process to kynurenine(GO:0019441) 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031251 | PAN complex(GO:0031251) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.3 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 1.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.1 | 0.5 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.1 | 0.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.3 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.4 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 1.1 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.0 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.8 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |