Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
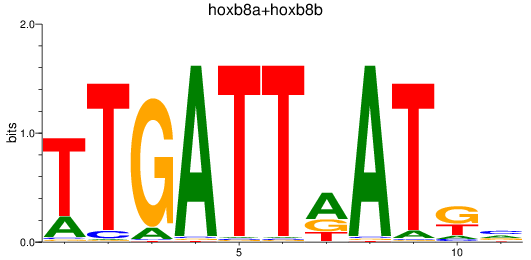
Results for hoxb8a+hoxb8b
Z-value: 0.74
Transcription factors associated with hoxb8a+hoxb8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb8b
|
ENSDARG00000054025 | homeobox B8b |
|
hoxb8a
|
ENSDARG00000056027 | homeobox B8a |
|
hoxb8b
|
ENSDARG00000111056 | homeobox B8b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb8b | dr11_v1_chr12_+_27117609_27117609 | 0.06 | 8.2e-01 | Click! |
| hoxb8a | dr11_v1_chr3_+_23687909_23687909 | 0.04 | 8.9e-01 | Click! |
Activity profile of hoxb8a+hoxb8b motif
Sorted Z-values of hoxb8a+hoxb8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_4461104 | 2.19 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr1_+_16144615 | 1.69 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr17_-_2595736 | 1.51 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr17_-_2573021 | 1.49 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr23_+_12545114 | 1.27 |
ENSDART00000105283
ENSDART00000166990 |
si:zfos-452g4.1
|
si:zfos-452g4.1 |
| chr11_+_13528437 | 1.03 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr11_+_3959495 | 0.92 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr21_-_31241254 | 0.91 |
ENSDART00000065365
|
tpst1l
|
tyrosylprotein sulfotransferase 1, like |
| chr24_-_7826489 | 0.86 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr16_-_26855936 | 0.84 |
ENSDART00000167320
ENSDART00000078119 |
ino80c
|
INO80 complex subunit C |
| chr3_+_26813058 | 0.80 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr7_+_12927944 | 0.78 |
ENSDART00000027329
|
fth1a
|
ferritin, heavy polypeptide 1a |
| chr2_-_10821053 | 0.75 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr17_-_2578026 | 0.71 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr8_-_49495584 | 0.68 |
ENSDART00000141691
|
opn7d
|
opsin 7, group member d |
| chr17_-_8570257 | 0.68 |
ENSDART00000154713
ENSDART00000121488 |
fzd3b
|
frizzled class receptor 3b |
| chr22_+_35089031 | 0.65 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr18_-_26715156 | 0.64 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr9_+_50000504 | 0.64 |
ENSDART00000164409
ENSDART00000165451 ENSDART00000166509 |
slc38a11
|
solute carrier family 38, member 11 |
| chr17_-_38778826 | 0.64 |
ENSDART00000168182
ENSDART00000124041 ENSDART00000136921 |
dglucy
|
D-glutamate cyclase |
| chr19_+_12444943 | 0.62 |
ENSDART00000135706
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr5_-_25733745 | 0.60 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr16_+_10776688 | 0.60 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr3_-_34717882 | 0.58 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr7_+_52154215 | 0.57 |
ENSDART00000098712
|
TMEM208
|
zgc:77041 |
| chr20_+_54274431 | 0.57 |
ENSDART00000099338
|
actr10
|
ARP10 actin related protein 10 homolog |
| chr9_-_9415000 | 0.56 |
ENSDART00000146210
|
si:ch211-214p13.9
|
si:ch211-214p13.9 |
| chr3_+_23752150 | 0.56 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr13_-_36621926 | 0.56 |
ENSDART00000057155
|
cdkn3
|
cyclin-dependent kinase inhibitor 3 |
| chr13_+_8604710 | 0.53 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr15_-_34567370 | 0.53 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr24_+_38301080 | 0.50 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr20_-_22778394 | 0.50 |
ENSDART00000152645
|
fip1l1a
|
FIP1 like 1a (S. cerevisiae) |
| chr3_+_33300522 | 0.49 |
ENSDART00000114023
|
hspb9
|
heat shock protein, alpha-crystallin-related, 9 |
| chr1_-_9277986 | 0.49 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr9_+_42066030 | 0.46 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr15_+_32405959 | 0.46 |
ENSDART00000177269
|
si:ch211-162k9.6
|
si:ch211-162k9.6 |
| chr9_-_43644261 | 0.46 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr5_-_34185115 | 0.45 |
ENSDART00000192771
|
fibcd1
|
fibrinogen C domain containing 1 |
| chr18_+_783936 | 0.44 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr11_-_3959477 | 0.42 |
ENSDART00000045971
|
pbrm1
|
polybromo 1 |
| chr10_-_39011514 | 0.42 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr24_+_34113424 | 0.42 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr6_-_53281518 | 0.40 |
ENSDART00000157621
|
rbm5
|
RNA binding motif protein 5 |
| chr1_-_19402802 | 0.39 |
ENSDART00000135552
|
rbm47
|
RNA binding motif protein 47 |
| chr5_-_825920 | 0.39 |
ENSDART00000126982
|
zgc:158463
|
zgc:158463 |
| chr4_-_9909371 | 0.39 |
ENSDART00000102656
|
si:dkey-22l11.6
|
si:dkey-22l11.6 |
| chr19_+_28256076 | 0.38 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr10_+_43039947 | 0.37 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr11_-_3959889 | 0.37 |
ENSDART00000159683
|
pbrm1
|
polybromo 1 |
| chr6_+_50451337 | 0.35 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr24_+_26328787 | 0.35 |
ENSDART00000003884
|
mynn
|
myoneurin |
| chr24_+_26329018 | 0.34 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr19_+_5480327 | 0.34 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr19_-_3876877 | 0.33 |
ENSDART00000163711
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr7_-_35126374 | 0.32 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr7_-_46777876 | 0.32 |
ENSDART00000193954
|
tshz3b
|
teashirt zinc finger homeobox 3b |
| chr3_+_59784632 | 0.32 |
ENSDART00000084729
|
pecam1
|
platelet/endothelial cell adhesion molecule 1 |
| chr24_+_5789790 | 0.31 |
ENSDART00000189600
|
BX470065.1
|
|
| chr24_-_6898302 | 0.31 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr2_+_7818368 | 0.30 |
ENSDART00000007068
|
kcnmb2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr18_-_26715655 | 0.30 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr12_+_17201522 | 0.30 |
ENSDART00000014536
ENSDART00000152452 |
rnls
|
renalase, FAD-dependent amine oxidase |
| chr2_+_3809226 | 0.30 |
ENSDART00000147261
|
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr7_+_21272833 | 0.30 |
ENSDART00000052942
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr3_-_13599482 | 0.29 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr12_-_28848015 | 0.29 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr1_-_43862638 | 0.29 |
ENSDART00000145044
|
tacr3a
|
tachykinin receptor 3a |
| chr24_+_5789582 | 0.29 |
ENSDART00000141504
|
BX470065.1
|
|
| chr10_-_45210184 | 0.29 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr6_-_23931442 | 0.29 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr2_-_24962820 | 0.28 |
ENSDART00000182767
|
hltf
|
helicase-like transcription factor |
| chr9_-_20372977 | 0.27 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr15_-_15968883 | 0.27 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr14_-_6666854 | 0.25 |
ENSDART00000133031
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr1_-_26071535 | 0.25 |
ENSDART00000185187
|
pdcd4a
|
programmed cell death 4a |
| chr7_+_13382852 | 0.25 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr10_+_40321067 | 0.25 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr11_+_36243774 | 0.24 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr21_-_23308286 | 0.24 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr4_-_5366566 | 0.22 |
ENSDART00000150788
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr18_-_14836862 | 0.22 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr15_+_8767650 | 0.22 |
ENSDART00000033871
|
ap2s1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr5_+_19261391 | 0.21 |
ENSDART00000089173
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr4_+_13810811 | 0.21 |
ENSDART00000067168
|
pdzrn4
|
PDZ domain containing ring finger 4 |
| chr14_+_5383060 | 0.21 |
ENSDART00000187825
|
lbx2
|
ladybird homeobox 2 |
| chr10_-_27741793 | 0.21 |
ENSDART00000129369
ENSDART00000192440 ENSDART00000189808 ENSDART00000138149 |
auts2a
si:dkey-33o22.1
|
autism susceptibility candidate 2a si:dkey-33o22.1 |
| chr5_+_19261687 | 0.20 |
ENSDART00000139401
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr16_+_17389116 | 0.19 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr16_-_54498109 | 0.19 |
ENSDART00000083713
|
clk2b
|
CDC-like kinase 2b |
| chr8_+_20503842 | 0.18 |
ENSDART00000100328
|
csnk1g2b
|
casein kinase 1, gamma 2b |
| chr2_+_47623202 | 0.17 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr12_+_39685485 | 0.17 |
ENSDART00000163403
|
LO017650.1
|
|
| chr4_+_7391110 | 0.17 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr24_+_25259154 | 0.16 |
ENSDART00000171125
|
gabrr3b
|
gamma-aminobutyric acid (GABA) A receptor, rho 3b |
| chr21_-_40348790 | 0.16 |
ENSDART00000178123
|
CR847523.1
|
|
| chr11_-_2838699 | 0.15 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr8_+_18588551 | 0.15 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr12_-_38548299 | 0.15 |
ENSDART00000153374
|
si:dkey-1f1.3
|
si:dkey-1f1.3 |
| chr13_+_22316746 | 0.15 |
ENSDART00000188968
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr25_+_15647750 | 0.15 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr4_-_55728559 | 0.14 |
ENSDART00000186201
|
CT583728.14
|
|
| chr8_+_21437908 | 0.14 |
ENSDART00000142758
|
si:dkey-163f12.10
|
si:dkey-163f12.10 |
| chr21_+_40092301 | 0.14 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr15_+_28202170 | 0.14 |
ENSDART00000077736
|
vtna
|
vitronectin a |
| chr25_+_15647993 | 0.14 |
ENSDART00000186578
ENSDART00000031828 |
spon1b
|
spondin 1b |
| chr12_-_11457625 | 0.14 |
ENSDART00000012318
|
htra1b
|
HtrA serine peptidase 1b |
| chr15_-_30450898 | 0.14 |
ENSDART00000156584
|
msi2b
|
musashi RNA-binding protein 2b |
| chr7_+_31879649 | 0.13 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr2_+_2223837 | 0.13 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr24_+_13735616 | 0.13 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr9_-_15789526 | 0.13 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr6_-_32087108 | 0.13 |
ENSDART00000150934
ENSDART00000130627 |
alg6
|
asparagine-linked glycosylation 6 (alpha-1,3,-glucosyltransferase) |
| chr17_+_39242437 | 0.13 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr20_-_19590378 | 0.13 |
ENSDART00000152588
|
baalcb
|
brain and acute leukemia, cytoplasmic b |
| chr19_-_3123963 | 0.13 |
ENSDART00000122816
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr1_+_51191049 | 0.13 |
ENSDART00000132244
ENSDART00000014970 ENSDART00000132141 |
btbd3a
|
BTB (POZ) domain containing 3a |
| chr13_-_5569562 | 0.12 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr11_+_6819050 | 0.12 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr18_-_38088099 | 0.12 |
ENSDART00000146120
|
luzp2
|
leucine zipper protein 2 |
| chr24_-_40009446 | 0.12 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr14_+_22297257 | 0.12 |
ENSDART00000113676
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr24_+_5840258 | 0.11 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr13_-_5568928 | 0.11 |
ENSDART00000192679
|
meis1b
|
Meis homeobox 1 b |
| chr13_+_36622100 | 0.11 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr25_-_16157776 | 0.11 |
ENSDART00000138453
|
si:dkey-80c24.5
|
si:dkey-80c24.5 |
| chr21_-_22812417 | 0.11 |
ENSDART00000079151
ENSDART00000112175 |
trpc6a
|
transient receptor potential cation channel, subfamily C, member 6a |
| chr23_+_21479958 | 0.11 |
ENSDART00000188302
ENSDART00000144320 |
si:dkey-1c11.1
|
si:dkey-1c11.1 |
| chr12_-_26406323 | 0.10 |
ENSDART00000131896
|
myoz1b
|
myozenin 1b |
| chr13_-_8776474 | 0.10 |
ENSDART00000122371
|
STPG4
|
si:ch211-93n23.7 |
| chr1_+_23784905 | 0.10 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr7_+_27976448 | 0.10 |
ENSDART00000181026
|
tub
|
tubby bipartite transcription factor |
| chr2_-_10188598 | 0.10 |
ENSDART00000189122
|
dmbx1a
|
diencephalon/mesencephalon homeobox 1a |
| chr22_+_18389271 | 0.10 |
ENSDART00000088270
|
yjefn3
|
YjeF N-terminal domain containing 3 |
| chr11_+_23265157 | 0.10 |
ENSDART00000110152
|
csf1a
|
colony stimulating factor 1a (macrophage) |
| chr24_-_26328721 | 0.09 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr15_+_28116129 | 0.09 |
ENSDART00000185157
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr7_-_31938938 | 0.09 |
ENSDART00000132353
|
bdnf
|
brain-derived neurotrophic factor |
| chr21_+_5192016 | 0.09 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr12_-_3756405 | 0.09 |
ENSDART00000150839
|
fam57bb
|
family with sequence similarity 57, member Bb |
| chr7_+_31879986 | 0.09 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr6_+_14949950 | 0.09 |
ENSDART00000149202
ENSDART00000149949 |
pou3f3b
|
POU class 3 homeobox 3b |
| chr8_-_14484599 | 0.08 |
ENSDART00000057644
|
lhx4
|
LIM homeobox 4 |
| chr1_-_8980665 | 0.08 |
ENSDART00000148182
|
si:ch73-191k20.3
|
si:ch73-191k20.3 |
| chr3_-_33574576 | 0.08 |
ENSDART00000184881
|
CR847537.1
|
|
| chr11_+_43419809 | 0.08 |
ENSDART00000172982
|
slc29a1b
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1b |
| chr13_-_30027730 | 0.08 |
ENSDART00000044009
|
scdb
|
stearoyl-CoA desaturase b |
| chr15_+_29727799 | 0.08 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr17_-_30666037 | 0.08 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr14_+_46274611 | 0.08 |
ENSDART00000134363
|
cabp2b
|
calcium binding protein 2b |
| chr4_-_77331797 | 0.08 |
ENSDART00000162325
|
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr3_-_29508959 | 0.08 |
ENSDART00000055408
|
cyth4a
|
cytohesin 4a |
| chr7_+_42460302 | 0.07 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr15_-_5467477 | 0.07 |
ENSDART00000123839
|
arrb1
|
arrestin, beta 1 |
| chr4_-_9891874 | 0.07 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr6_+_52804267 | 0.07 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr20_-_8443425 | 0.07 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr5_-_24869213 | 0.07 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr5_+_9382301 | 0.07 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr20_-_29420713 | 0.07 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr20_-_43663494 | 0.06 |
ENSDART00000144564
|
BX470188.1
|
|
| chr14_+_25465346 | 0.06 |
ENSDART00000173436
|
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr7_-_69983948 | 0.06 |
ENSDART00000185827
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr1_-_21901589 | 0.06 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr21_+_22423286 | 0.06 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr14_+_4796168 | 0.06 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr20_-_32188897 | 0.06 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr2_-_54039293 | 0.06 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr13_+_30912385 | 0.06 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr4_-_77332032 | 0.06 |
ENSDART00000168628
ENSDART00000172025 |
slco1f3
|
solute carrier organic anion transporter family, member 1F3 |
| chr21_-_27338639 | 0.06 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr1_-_47431453 | 0.06 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr1_+_57151926 | 0.06 |
ENSDART00000152628
|
si:ch73-94k4.5
|
si:ch73-94k4.5 |
| chr2_-_9646857 | 0.06 |
ENSDART00000056901
|
zgc:153615
|
zgc:153615 |
| chr24_+_24461558 | 0.06 |
ENSDART00000182424
|
bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr1_+_56801021 | 0.05 |
ENSDART00000146545
|
si:ch211-1f22.3
|
si:ch211-1f22.3 |
| chr25_+_5972690 | 0.05 |
ENSDART00000067517
|
si:ch211-11i22.4
|
si:ch211-11i22.4 |
| chr2_+_38608290 | 0.05 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr9_-_3934963 | 0.05 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr20_-_40754794 | 0.05 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr5_+_19094462 | 0.05 |
ENSDART00000190596
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr3_+_18936009 | 0.05 |
ENSDART00000140330
|
si:ch211-198m1.1
|
si:ch211-198m1.1 |
| chr11_+_3281899 | 0.05 |
ENSDART00000181012
|
mmp19
|
matrix metallopeptidase 19 |
| chr24_+_41915878 | 0.05 |
ENSDART00000171523
|
TMEM200C
|
transmembrane protein 200C |
| chr4_+_20954929 | 0.05 |
ENSDART00000143674
|
nav3
|
neuron navigator 3 |
| chr20_-_2949028 | 0.05 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr23_-_7674902 | 0.05 |
ENSDART00000185612
ENSDART00000180524 |
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr10_-_8197049 | 0.04 |
ENSDART00000129467
|
dhx29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr1_+_57208005 | 0.04 |
ENSDART00000152410
|
si:dkey-27j5.10
|
si:dkey-27j5.10 |
| chr15_-_21132480 | 0.04 |
ENSDART00000078734
ENSDART00000157481 |
a2ml
|
alpha-2-macroglobulin-like |
| chr23_+_5490854 | 0.04 |
ENSDART00000175403
|
tulp1a
|
tubby like protein 1a |
| chr23_-_11870962 | 0.04 |
ENSDART00000143481
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr7_+_37377335 | 0.04 |
ENSDART00000111842
|
sall1a
|
spalt-like transcription factor 1a |
| chr10_-_7785930 | 0.04 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr6_-_43677125 | 0.04 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr20_-_26491567 | 0.04 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr6_-_18960105 | 0.04 |
ENSDART00000185278
ENSDART00000162968 |
sept9b
|
septin 9b |
| chr1_+_26071869 | 0.04 |
ENSDART00000059264
|
mxd4
|
MAX dimerization protein 4 |
| chr24_-_28893251 | 0.04 |
ENSDART00000042065
ENSDART00000003503 |
col11a1a
|
collagen, type XI, alpha 1a |
| chr4_+_54618332 | 0.04 |
ENSDART00000171824
|
si:ch211-227e10.2
|
si:ch211-227e10.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb8a+hoxb8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 3.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.8 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.9 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.8 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.9 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 2.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.4 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.2 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.6 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.3 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.0 | 0.2 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.5 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) potassium ion homeostasis(GO:0055075) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 1.3 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.6 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.2 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.6 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.7 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.7 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.5 | GO:0007259 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 0.1 | GO:2001287 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.6 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.5 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.5 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 1.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 2.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0030130 | trans-Golgi network transport vesicle membrane(GO:0012510) clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 2.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.9 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.3 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.8 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:1902388 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.7 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.7 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.8 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |