Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
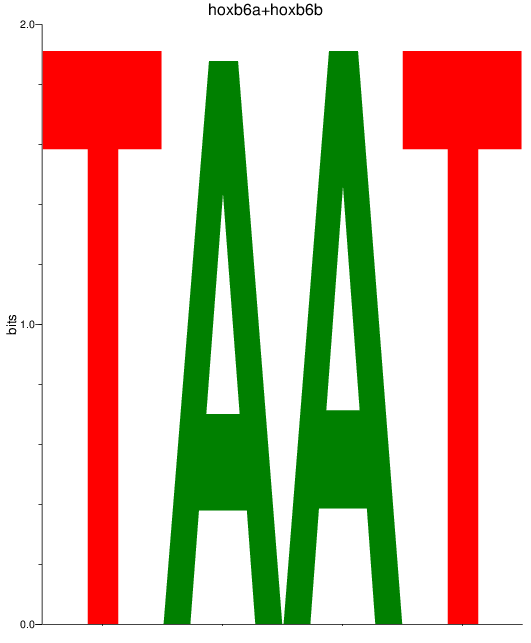
Results for hoxb6a+hoxb6b
Z-value: 1.67
Transcription factors associated with hoxb6a+hoxb6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb6a
|
ENSDARG00000010630 | homeobox B6a |
|
hoxb6b
|
ENSDARG00000026513 | homeobox B6b |
|
hoxb6b
|
ENSDARG00000111786 | homeobox B6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb6a | dr11_v1_chr3_+_23703704_23703704 | -0.54 | 2.0e-02 | Click! |
| hoxb6b | dr11_v1_chr12_+_27127139_27127139 | -0.22 | 3.8e-01 | Click! |
Activity profile of hoxb6a+hoxb6b motif
Sorted Z-values of hoxb6a+hoxb6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_24717365 | 3.47 |
ENSDART00000137934
ENSDART00000003922 |
erlin1
|
ER lipid raft associated 1 |
| chr13_-_24717618 | 3.42 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr20_-_38787047 | 2.84 |
ENSDART00000152913
ENSDART00000153430 |
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr7_+_41812190 | 2.50 |
ENSDART00000113732
ENSDART00000174137 |
orc6
|
origin recognition complex, subunit 6 |
| chr4_-_5775507 | 2.48 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr5_-_9793273 | 2.46 |
ENSDART00000158874
|
ddr2l
|
discoidin domain receptor family, member 2, like |
| chr20_-_38787341 | 2.44 |
ENSDART00000136771
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr21_-_19918286 | 2.44 |
ENSDART00000180816
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr22_+_11756040 | 2.38 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr17_+_16090436 | 2.29 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr19_+_2835240 | 2.21 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr7_+_33424044 | 2.17 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr1_-_33647138 | 2.16 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr6_+_4528631 | 2.14 |
ENSDART00000122042
|
RNF219
|
ring finger protein 219 |
| chr3_-_40054615 | 2.13 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr25_-_7918526 | 2.09 |
ENSDART00000104686
ENSDART00000156761 |
ambra1b
|
autophagy/beclin-1 regulator 1b |
| chr11_+_31121340 | 2.04 |
ENSDART00000185172
|
stx10
|
syntaxin 10 |
| chr19_-_4137087 | 2.01 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr13_-_36761379 | 2.01 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr25_-_29074064 | 2.00 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr4_+_5341592 | 1.97 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr20_-_38778479 | 1.94 |
ENSDART00000185599
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr19_-_30800004 | 1.92 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr6_-_7107868 | 1.90 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr23_+_19590598 | 1.89 |
ENSDART00000170149
|
slmapb
|
sarcolemma associated protein b |
| chr15_+_28175638 | 1.87 |
ENSDART00000037119
|
slc46a1
|
solute carrier family 46 (folate transporter), member 1 |
| chr15_+_2190229 | 1.84 |
ENSDART00000147710
|
rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr24_-_2450597 | 1.83 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr2_-_17115256 | 1.82 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr11_-_43200994 | 1.80 |
ENSDART00000164700
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_+_29794058 | 1.79 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr7_+_27603211 | 1.74 |
ENSDART00000148782
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr8_-_1838315 | 1.74 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr17_+_11372531 | 1.74 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr12_-_33357655 | 1.74 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_-_19919020 | 1.72 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr5_-_39171302 | 1.72 |
ENSDART00000020808
|
paqr3a
|
progestin and adipoQ receptor family member IIIa |
| chr11_-_27537593 | 1.70 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr16_-_54919260 | 1.69 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr16_-_41646164 | 1.69 |
ENSDART00000184257
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr5_-_68333081 | 1.69 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr12_-_13730501 | 1.68 |
ENSDART00000152370
|
foxh1
|
forkhead box H1 |
| chr16_+_38940758 | 1.67 |
ENSDART00000102482
ENSDART00000136215 |
eny2
|
enhancer of yellow 2 homolog (Drosophila) |
| chr7_+_69187585 | 1.66 |
ENSDART00000160499
ENSDART00000166258 |
marveld3
|
MARVEL domain containing 3 |
| chr18_+_6638974 | 1.66 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr16_+_16265850 | 1.66 |
ENSDART00000181265
|
setd2
|
SET domain containing 2 |
| chr17_-_23895026 | 1.65 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr16_+_40043673 | 1.65 |
ENSDART00000102552
ENSDART00000125484 |
trmt11
|
tRNA methyltransferase 11 homolog (S. cerevisiae) |
| chr7_-_33683891 | 1.64 |
ENSDART00000175980
ENSDART00000191148 ENSDART00000173569 |
tle3b
|
transducin-like enhancer of split 3b |
| chr14_+_22132388 | 1.63 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr17_-_41798856 | 1.62 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr3_+_7808459 | 1.61 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr15_-_28908027 | 1.61 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr20_-_23253630 | 1.61 |
ENSDART00000103365
|
ociad1
|
OCIA domain containing 1 |
| chr6_-_40713183 | 1.60 |
ENSDART00000157113
ENSDART00000154810 ENSDART00000153702 |
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr18_-_35407695 | 1.59 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr23_-_43595956 | 1.58 |
ENSDART00000162186
|
itchb
|
itchy E3 ubiquitin protein ligase b |
| chr18_+_3243292 | 1.58 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_+_7703251 | 1.56 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr11_-_44979281 | 1.56 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr1_-_55044256 | 1.55 |
ENSDART00000165505
ENSDART00000167536 ENSDART00000170001 |
vps54
|
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr17_+_17764979 | 1.53 |
ENSDART00000105013
|
alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr6_-_32045951 | 1.53 |
ENSDART00000016629
ENSDART00000139055 |
efcab7
|
EF-hand calcium binding domain 7 |
| chr14_+_989733 | 1.52 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr5_+_31214341 | 1.52 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr2_-_32237916 | 1.51 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr16_-_54907588 | 1.51 |
ENSDART00000185709
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr14_+_28492990 | 1.51 |
ENSDART00000160347
ENSDART00000187176 ENSDART00000088094 |
stag2b
|
stromal antigen 2b |
| chr18_-_35407530 | 1.50 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr14_+_34492288 | 1.49 |
ENSDART00000144301
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr13_+_21674445 | 1.48 |
ENSDART00000100931
ENSDART00000078567 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr5_+_25733774 | 1.48 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr17_-_45125537 | 1.47 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr16_-_39267185 | 1.46 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr1_-_23293261 | 1.46 |
ENSDART00000122648
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr12_+_22407852 | 1.45 |
ENSDART00000178840
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr17_+_16192486 | 1.44 |
ENSDART00000156832
|
kif13ba
|
kinesin family member 13Ba |
| chr13_-_24825691 | 1.44 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr24_-_26622423 | 1.44 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr9_-_54840124 | 1.43 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr10_-_21362320 | 1.43 |
ENSDART00000189789
|
avd
|
avidin |
| chr7_-_45852270 | 1.42 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr16_-_29387215 | 1.42 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr12_-_10508952 | 1.42 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr7_+_60396957 | 1.42 |
ENSDART00000121545
|
brms1
|
breast cancer metastasis suppressor 1 |
| chr21_-_13856689 | 1.41 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr7_+_41812636 | 1.41 |
ENSDART00000174333
|
orc6
|
origin recognition complex, subunit 6 |
| chr14_+_21699129 | 1.40 |
ENSDART00000073707
|
stx3a
|
syntaxin 3A |
| chr3_+_35542067 | 1.40 |
ENSDART00000146529
ENSDART00000084549 |
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr14_-_34605607 | 1.40 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr9_-_12401871 | 1.40 |
ENSDART00000191901
|
nup35
|
nucleoporin 35 |
| chr17_-_30635298 | 1.40 |
ENSDART00000155478
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr12_+_30788912 | 1.39 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr14_-_31854830 | 1.39 |
ENSDART00000148550
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr4_-_27099224 | 1.39 |
ENSDART00000048383
|
creld2
|
cysteine-rich with EGF-like domains 2 |
| chr8_+_47677208 | 1.38 |
ENSDART00000123254
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr18_+_6638726 | 1.38 |
ENSDART00000142755
ENSDART00000167781 |
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr17_-_4252221 | 1.38 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr15_+_12429206 | 1.38 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr19_+_7549854 | 1.37 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr11_-_2504750 | 1.37 |
ENSDART00000173149
|
dgkab
|
diacylglycerol kinase, alpha b |
| chr19_-_24136233 | 1.37 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr17_+_19626479 | 1.36 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr14_-_41308561 | 1.36 |
ENSDART00000138232
|
arl13a
|
ADP-ribosylation factor-like 13A |
| chr1_-_26664840 | 1.35 |
ENSDART00000102500
|
anp32b
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
| chr18_+_6641542 | 1.35 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr6_+_20954400 | 1.35 |
ENSDART00000143248
ENSDART00000165806 |
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr7_-_71585065 | 1.35 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr17_-_31483469 | 1.34 |
ENSDART00000062907
ENSDART00000061547 |
ltk
|
leukocyte receptor tyrosine kinase |
| chr6_-_11362871 | 1.34 |
ENSDART00000151125
|
pcnt
|
pericentrin |
| chr6_-_7109063 | 1.33 |
ENSDART00000148548
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr8_+_11471350 | 1.33 |
ENSDART00000092355
ENSDART00000136184 |
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr13_-_24257631 | 1.33 |
ENSDART00000146524
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr5_-_15494164 | 1.32 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr3_-_30885250 | 1.32 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr21_+_15870752 | 1.32 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr19_-_2115040 | 1.32 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr25_-_25058508 | 1.32 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr13_+_30035253 | 1.32 |
ENSDART00000181303
ENSDART00000057525 ENSDART00000136622 |
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr18_-_35407289 | 1.32 |
ENSDART00000012018
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr5_+_13521081 | 1.32 |
ENSDART00000171975
|
si:ch211-230g14.6
|
si:ch211-230g14.6 |
| chr16_+_30117798 | 1.31 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr3_+_40409100 | 1.31 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr12_+_22672323 | 1.31 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr1_-_23370395 | 1.31 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr14_+_35414632 | 1.31 |
ENSDART00000191516
ENSDART00000084914 |
trmt12
|
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr24_+_41989108 | 1.31 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr10_-_3427589 | 1.30 |
ENSDART00000133452
ENSDART00000037183 |
tmed2
|
transmembrane p24 trafficking protein 2 |
| chr1_-_6028876 | 1.30 |
ENSDART00000168117
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr8_+_28452738 | 1.30 |
ENSDART00000062706
|
tmem189
|
transmembrane protein 189 |
| chr13_+_25505580 | 1.29 |
ENSDART00000140634
|
inpp5f
|
inositol polyphosphate-5-phosphatase F |
| chr8_+_11425048 | 1.29 |
ENSDART00000018739
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr16_+_30002605 | 1.29 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr8_+_2543525 | 1.29 |
ENSDART00000129569
ENSDART00000128227 |
gle1
|
GLE1 RNA export mediator homolog (yeast) |
| chr5_-_6567464 | 1.29 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr7_+_28724919 | 1.29 |
ENSDART00000011324
|
ccdc102a
|
coiled-coil domain containing 102A |
| chr5_+_8196264 | 1.29 |
ENSDART00000174564
ENSDART00000161261 |
lmbrd2a
|
LMBR1 domain containing 2a |
| chr1_-_43920371 | 1.28 |
ENSDART00000109283
|
scpp7
|
secretory calcium-binding phosphoprotein 7 |
| chr12_+_22628704 | 1.28 |
ENSDART00000168935
|
mfsd8
|
major facilitator superfamily domain containing 8 |
| chr20_-_14781904 | 1.28 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr14_-_26498196 | 1.28 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr10_-_21362071 | 1.28 |
ENSDART00000125167
|
avd
|
avidin |
| chr2_+_15048410 | 1.28 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr2_-_37477654 | 1.27 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr24_-_16979728 | 1.27 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr22_-_22337382 | 1.27 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr11_-_35171768 | 1.26 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr7_+_36470159 | 1.26 |
ENSDART00000188703
|
aktip
|
akt interacting protein |
| chr3_+_54047342 | 1.26 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr13_+_7665890 | 1.26 |
ENSDART00000046792
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr9_+_23003208 | 1.26 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr2_-_54387550 | 1.26 |
ENSDART00000097388
|
napgb
|
N-ethylmaleimide-sensitive factor attachment protein, gamma b |
| chr17_+_51744450 | 1.25 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr8_-_16725573 | 1.24 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr2_-_55779927 | 1.24 |
ENSDART00000168579
|
CABZ01066725.1
|
|
| chr7_+_32695954 | 1.23 |
ENSDART00000184425
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr20_+_33924235 | 1.23 |
ENSDART00000146292
ENSDART00000139609 |
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr18_+_18000695 | 1.23 |
ENSDART00000146898
|
si:ch211-212o1.2
|
si:ch211-212o1.2 |
| chr1_-_52790724 | 1.23 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr2_-_37103622 | 1.23 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr3_+_27606505 | 1.22 |
ENSDART00000150953
|
usp7
|
ubiquitin specific peptidase 7 (herpes virus-associated) |
| chr3_-_26184018 | 1.22 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr14_+_34490445 | 1.22 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr22_+_15960514 | 1.22 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr13_-_38730267 | 1.22 |
ENSDART00000157524
|
lmbrd1
|
LMBR1 domain containing 1 |
| chr8_-_20230559 | 1.22 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr6_+_29693492 | 1.21 |
ENSDART00000114172
|
pde6d
|
phosphodiesterase 6D, cGMP-specific, rod, delta |
| chr16_+_16266428 | 1.21 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr20_-_43750771 | 1.21 |
ENSDART00000100605
|
ttc32
|
tetratricopeptide repeat domain 32 |
| chr4_-_15003854 | 1.21 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr16_+_20056030 | 1.21 |
ENSDART00000027020
|
ankrd28b
|
ankyrin repeat domain 28b |
| chr3_-_30488063 | 1.20 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr4_+_27898833 | 1.20 |
ENSDART00000146099
|
cerk
|
ceramide kinase |
| chr14_+_45028062 | 1.20 |
ENSDART00000184717
ENSDART00000185481 |
ATP8A1
|
ATPase phospholipid transporting 8A1 |
| chr5_+_41477954 | 1.20 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr5_-_54481692 | 1.19 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr8_-_49935358 | 1.19 |
ENSDART00000159782
ENSDART00000156841 |
agtpbp1
CU694202.1
|
ATP/GTP binding protein 1 |
| chr17_+_30591287 | 1.19 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr23_+_2704793 | 1.18 |
ENSDART00000147953
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr7_+_22792132 | 1.18 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr7_-_64971839 | 1.18 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr25_+_17689565 | 1.18 |
ENSDART00000171965
|
galnt18a
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18a |
| chr22_-_881725 | 1.18 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr19_-_11846958 | 1.18 |
ENSDART00000148516
|
ctdp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr22_-_5171829 | 1.18 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr20_-_48898560 | 1.18 |
ENSDART00000163071
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr1_+_46493944 | 1.18 |
ENSDART00000114083
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr25_+_3759553 | 1.17 |
ENSDART00000180601
ENSDART00000055845 ENSDART00000157050 ENSDART00000153905 |
thoc5
|
THO complex 5 |
| chr12_+_14149686 | 1.17 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr5_-_10082244 | 1.17 |
ENSDART00000036421
|
chek2
|
checkpoint kinase 2 |
| chr11_-_10456553 | 1.16 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr1_+_12394205 | 1.16 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr21_+_18405585 | 1.16 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr19_+_29808471 | 1.16 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr23_+_2789422 | 1.16 |
ENSDART00000156954
|
plcg1
|
phospholipase C, gamma 1 |
| chr13_+_29926631 | 1.16 |
ENSDART00000135265
|
cuedc2
|
CUE domain containing 2 |
| chr20_-_3319642 | 1.16 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr25_-_2723682 | 1.15 |
ENSDART00000113382
|
adpgk
|
ADP-dependent glucokinase |
| chr21_-_34032650 | 1.15 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr16_+_20055878 | 1.15 |
ENSDART00000146436
|
ankrd28b
|
ankyrin repeat domain 28b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb6a+hoxb6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 1.1 | 3.3 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.8 | 3.8 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.6 | 2.5 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.6 | 3.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.6 | 1.8 | GO:0010660 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.6 | 4.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.5 | 7.0 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.5 | 2.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.5 | 2.0 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 2.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.5 | 1.4 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.5 | 2.9 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.5 | 1.4 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.5 | 1.4 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.5 | 1.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 2.7 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 | 1.3 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.4 | 1.3 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 2.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 3.0 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.4 | 1.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.4 | 1.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.4 | 1.2 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.4 | 1.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.4 | 1.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.4 | 1.5 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.1 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.4 | 2.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.3 | 2.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.3 | 1.0 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.3 | 1.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 1.7 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.3 | 1.9 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.3 | 1.9 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.3 | 1.5 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.3 | 0.9 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.3 | 2.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.3 | 1.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 1.7 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.3 | 1.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.3 | 0.8 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.3 | 0.8 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.3 | 1.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.3 | 1.1 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.3 | 2.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 1.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.3 | 1.8 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.3 | 0.8 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.3 | 0.5 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) |
| 0.3 | 1.8 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.3 | 1.3 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.3 | 0.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 1.0 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.7 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 2.9 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 1.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.2 | 0.9 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 0.9 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 1.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 0.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.7 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.2 | 0.6 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.2 | 0.6 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.2 | 0.6 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 2.4 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.6 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 0.8 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 0.6 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.2 | 2.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.2 | 4.3 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.2 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.9 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 2.0 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 0.9 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 1.5 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.2 | 0.7 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.2 | 1.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.2 | 2.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 | 0.9 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 1.6 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.2 | 0.5 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 1.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.2 | 1.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.2 | 1.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 3.9 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.2 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.2 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 2.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.2 | 0.8 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.2 | 0.8 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 3.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 1.4 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 0.8 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.2 | 0.8 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.9 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 1.3 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.7 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 2.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 1.3 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 1.4 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.3 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.2 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.8 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.8 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 1.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 0.7 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.9 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 1.0 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.1 | 1.0 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.5 | GO:0044090 | positive regulation of vacuole organization(GO:0044090) |
| 0.1 | 3.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.1 | 1.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 1.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 2.5 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 1.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.6 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 2.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.1 | 0.7 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.8 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 3.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.4 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 3.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.5 | GO:0016237 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.0 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.1 | 0.2 | GO:0042706 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) neuron fate determination(GO:0048664) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 2.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.4 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.1 | 1.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 1.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.4 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.4 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 1.1 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.6 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.1 | 1.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.7 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.9 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.9 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.3 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.3 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.7 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.1 | 0.6 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.1 | 0.8 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 2.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 1.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.7 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 0.7 | GO:0009146 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.7 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.6 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.9 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.5 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 0.5 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 1.2 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.1 | 0.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.6 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 0.6 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 2.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 2.2 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.8 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.1 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.6 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.1 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.6 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 1.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 1.5 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 2.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 1.4 | GO:0005979 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 0.4 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 1.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.4 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 1.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 0.5 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 0.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 3.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 2.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 1.4 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 1.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.3 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.1 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 2.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.0 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 0.9 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 0.4 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 1.9 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.5 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 2.9 | GO:0051170 | nuclear import(GO:0051170) |
| 0.1 | 1.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.1 | 0.6 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.0 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.9 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.2 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.1 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.6 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.1 | 0.5 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 0.2 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 2.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 2.4 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.6 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.1 | 0.6 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.5 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 0.7 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.3 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.5 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.1 | 5.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 1.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.2 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.2 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.1 | 0.4 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.9 | GO:0042770 | signal transduction in response to DNA damage(GO:0042770) |
| 0.1 | 0.4 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.2 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 2.8 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.2 | GO:0014060 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) positive regulation of amine transport(GO:0051954) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.7 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 2.0 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.4 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.8 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.9 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 1.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.7 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.5 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.0 | 0.7 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 2.3 | GO:0005977 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 4.1 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 1.2 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.0 | 1.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.4 | GO:0003159 | morphogenesis of an endothelium(GO:0003159) |
| 0.0 | 0.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 11.5 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.7 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.5 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.6 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.4 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.5 | GO:0002029 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.3 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 1.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.4 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 2.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 2.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.6 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.4 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.3 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.5 | GO:0039014 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) |
| 0.0 | 1.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.2 | GO:0034552 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.4 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.3 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 1.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 1.7 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.0 | 0.7 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.0 | 0.5 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0034354 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 1.0 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.4 | GO:0055008 | cardiac muscle tissue morphogenesis(GO:0055008) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.5 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.6 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 2.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 1.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.8 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.7 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 1.1 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.7 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.7 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.0 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.5 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0002823 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.5 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.0 | GO:0071027 | tRNA catabolic process(GO:0016078) U1 snRNA 3'-end processing(GO:0034473) U4 snRNA 3'-end processing(GO:0034475) U5 snRNA 3'-end processing(GO:0034476) polyadenylation-dependent RNA catabolic process(GO:0043633) polyadenylation-dependent ncRNA catabolic process(GO:0043634) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) nuclear ncRNA surveillance(GO:0071029) nuclear polyadenylation-dependent rRNA catabolic process(GO:0071035) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) nuclear polyadenylation-dependent ncRNA catabolic process(GO:0071046) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.0 | 0.1 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.4 | GO:0006623 | protein targeting to vacuole(GO:0006623) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 1.1 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.7 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.0 | 0.3 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0051341 | regulation of oxidoreductase activity(GO:0051341) |
| 0.0 | 0.3 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 2.4 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 1.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.8 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.0 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.5 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.6 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.7 | GO:0006672 | ceramide metabolic process(GO:0006672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.7 | 3.6 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.6 | 3.0 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.5 | 1.6 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.5 | 2.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.5 | 1.4 | GO:0044279 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.5 | 1.9 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.5 | 1.8 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.4 | 1.7 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.4 | 1.6 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.4 | 1.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.4 | 2.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.4 | 1.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.4 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 3.9 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 7.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 5.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.3 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.3 | 1.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 0.8 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 2.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 1.6 | GO:0000938 | GARP complex(GO:0000938) |
| 0.2 | 2.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 1.3 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 1.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 3.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 1.8 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.8 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 1.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 0.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 0.7 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.2 | 1.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.2 | 1.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 1.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.2 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 1.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 1.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.4 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 2.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.9 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 3.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 1.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 5.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 2.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 14.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 1.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 2.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.8 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.7 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.3 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.6 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.1 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.3 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.6 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 6.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 5.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 1.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.6 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.6 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 2.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 5.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 2.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 5.3 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.8 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 3.0 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.7 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.0 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 1.8 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.4 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 2.3 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.1 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.0 | 2.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.1 | GO:0070938 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 4.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.7 | 2.7 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.7 | 2.7 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.6 | 5.0 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.6 | 2.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.5 | 2.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.5 | 2.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.5 | 1.4 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.4 | 1.3 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.4 | 1.3 | GO:0052833 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.4 | 1.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.4 | 1.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.4 | 1.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.4 | 1.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.4 | 1.5 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.4 | 1.9 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.4 | 1.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 1.5 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.3 | 5.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.0 | GO:0042806 | fucose binding(GO:0042806) |
| 0.3 | 1.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 1.6 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 2.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 2.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.3 | 0.9 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 0.8 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.3 | 2.5 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 1.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.6 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 1.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 0.8 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.2 | 1.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 1.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 1.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.7 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.2 | 2.0 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.2 | 1.1 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 0.9 | GO:0033745 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 0.8 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.2 | 4.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.2 | 0.8 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.2 | 1.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 0.8 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.2 | 2.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.3 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.2 | 0.9 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 1.8 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.2 | 1.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 0.9 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.2 | 2.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.2 | 0.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.2 | 1.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 2.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 0.8 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.2 | 0.8 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 3.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 2.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.4 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 3.2 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.2 | 0.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 3.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 2.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 2.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 2.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 2.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 0.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 3.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 7.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 2.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 1.1 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.6 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.3 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 1.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.7 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.9 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.4 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 2.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.3 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.5 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 1.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 1.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 2.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.7 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 0.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.4 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 0.5 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.5 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 1.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.8 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 0.4 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 2.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.9 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 0.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 3.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.7 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.5 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.7 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.1 | GO:0035004 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.1 | 0.4 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 5.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.3 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 0.4 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 2.2 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.4 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.3 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 1.7 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 4.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 2.1 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 3.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.2 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 1.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 0.2 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) |
| 0.1 | 0.5 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 0.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 2.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.1 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.8 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.1 | 0.3 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 4.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.2 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.6 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 4.1 | GO:0043566 | structure-specific DNA binding(GO:0043566) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 5.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 1.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.9 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 1.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.1 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.0 | 3.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.6 | GO:0008144 | drug binding(GO:0008144) cyclosporin A binding(GO:0016018) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.6 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.2 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 2.1 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 1.0 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 5.3 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 8.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 4.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 6.8 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 5.1 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 2.6 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.1 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 2.3 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0016893 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) endonuclease activity, active with either ribo- or deoxyribonucleic acids and producing 5'-phosphomonoesters(GO:0016893) |
| 0.0 | 1.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0044389 | ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 2.4 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.4 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.0 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 4.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 1.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 2.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.2 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.2 | 1.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 5.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 3.0 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.8 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.1 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 5.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.1 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.1 | 1.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 2.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.6 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 2.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.0 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 0.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 0.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.9 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.4 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.3 | 2.7 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.3 | 1.2 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |
| 0.3 | 1.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 2.9 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 3.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 3.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 1.6 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.2 | 1.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.2 | 1.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.2 | 1.8 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 3.6 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 4.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.1 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.8 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 2.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.6 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.0 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.1 | 1.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.1 | 2.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.7 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 2.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 0.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.2 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.5 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.0 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.1 | 1.0 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.7 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 2.1 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 1.3 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.7 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 1.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |