Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
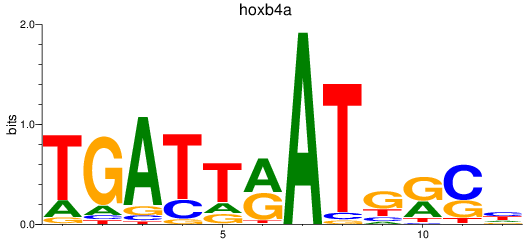
Results for hoxb4a
Z-value: 0.29
Transcription factors associated with hoxb4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb4a
|
ENSDARG00000013533 | homeobox B4a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb4a | dr11_v1_chr3_+_23710839_23710839 | -0.10 | 6.9e-01 | Click! |
Activity profile of hoxb4a motif
Sorted Z-values of hoxb4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_21406769 | 0.74 |
ENSDART00000135766
|
si:dkey-163f12.6
|
si:dkey-163f12.6 |
| chr19_+_816208 | 0.73 |
ENSDART00000093304
|
nrm
|
nurim |
| chr10_+_36026576 | 0.68 |
ENSDART00000193786
|
hmgb1a
|
high mobility group box 1a |
| chr21_+_22423286 | 0.62 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr17_+_23937262 | 0.61 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr25_-_4482449 | 0.60 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr5_+_20112032 | 0.54 |
ENSDART00000130554
|
isg15
|
ISG15 ubiquitin-like modifier |
| chr6_+_29791164 | 0.50 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr12_+_28854410 | 0.42 |
ENSDART00000152991
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr20_+_51478939 | 0.37 |
ENSDART00000149758
|
tlr5a
|
toll-like receptor 5a |
| chr5_+_32924669 | 0.34 |
ENSDART00000085219
|
lmo4a
|
LIM domain only 4a |
| chr8_-_912821 | 0.33 |
ENSDART00000082296
|
ptger4a
|
prostaglandin E receptor 4 (subtype EP4) a |
| chr1_-_46981134 | 0.32 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr17_-_19626357 | 0.32 |
ENSDART00000011432
|
reep3a
|
receptor accessory protein 3a |
| chr21_-_24632778 | 0.29 |
ENSDART00000132533
ENSDART00000058370 |
arhgap32b
|
Rho GTPase activating protein 32b |
| chr25_-_13842618 | 0.28 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr20_+_51479263 | 0.25 |
ENSDART00000148798
|
tlr5a
|
toll-like receptor 5a |
| chr10_-_26744131 | 0.23 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr21_-_27338639 | 0.22 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr9_+_45605410 | 0.19 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr12_+_28854963 | 0.19 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr3_+_27027781 | 0.17 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr23_+_3721042 | 0.17 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr3_-_51109286 | 0.17 |
ENSDART00000172010
|
si:ch211-148f13.1
|
si:ch211-148f13.1 |
| chr4_+_12340655 | 0.15 |
ENSDART00000168487
|
pimr214
|
Pim proto-oncogene, serine/threonine kinase, related 214 |
| chr9_-_15789526 | 0.15 |
ENSDART00000141318
|
si:dkey-103d23.3
|
si:dkey-103d23.3 |
| chr6_-_28345002 | 0.15 |
ENSDART00000158955
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr20_-_32188897 | 0.14 |
ENSDART00000133887
|
si:ch211-51a19.5
|
si:ch211-51a19.5 |
| chr7_-_20453661 | 0.14 |
ENSDART00000174001
|
ntn5
|
netrin 5 |
| chr15_-_15968883 | 0.14 |
ENSDART00000166583
ENSDART00000154042 |
synrg
|
synergin, gamma |
| chr5_-_7834582 | 0.12 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr4_+_7391110 | 0.12 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr20_-_29420713 | 0.12 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr20_-_26491567 | 0.11 |
ENSDART00000147154
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr7_-_52842007 | 0.11 |
ENSDART00000182710
|
map1aa
|
microtubule-associated protein 1Aa |
| chr13_+_36622100 | 0.10 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr17_-_114121 | 0.09 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr14_+_46274611 | 0.09 |
ENSDART00000134363
|
cabp2b
|
calcium binding protein 2b |
| chr17_-_23241393 | 0.08 |
ENSDART00000190697
|
AL935174.4
|
|
| chr17_+_39242437 | 0.08 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr6_-_29105727 | 0.07 |
ENSDART00000184355
|
fam69ab
|
family with sequence similarity 69, member Ab |
| chr15_+_17251191 | 0.07 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr18_-_14836862 | 0.07 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr18_-_14836600 | 0.07 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr17_+_27434626 | 0.07 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr2_-_722156 | 0.06 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr3_-_52644737 | 0.06 |
ENSDART00000126180
|
si:dkey-210j14.3
|
si:dkey-210j14.3 |
| chr7_+_35268880 | 0.04 |
ENSDART00000182231
|
dpep2
|
dipeptidase 2 |
| chr1_-_47431453 | 0.04 |
ENSDART00000101104
|
gja5b
|
gap junction protein, alpha 5b |
| chr1_+_11881559 | 0.04 |
ENSDART00000166981
|
snx8b
|
sorting nexin 8b |
| chr11_+_40530748 | 0.04 |
ENSDART00000173239
|
mmel1
|
membrane metallo-endopeptidase-like 1 |
| chr7_+_27317174 | 0.03 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr13_+_30912117 | 0.03 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr9_+_22351443 | 0.03 |
ENSDART00000080054
|
crygs3
|
crystallin, gamma S3 |
| chr12_+_9703172 | 0.03 |
ENSDART00000091489
|
ppp1r9bb
|
protein phosphatase 1, regulatory subunit 9Bb |
| chr13_+_30912385 | 0.02 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr10_-_20445549 | 0.02 |
ENSDART00000064613
|
loxl2a
|
lysyl oxidase-like 2a |
| chr24_-_26460853 | 0.02 |
ENSDART00000078628
|
slc7a14b
|
solute carrier family 7, member 14b |
| chr23_-_3721444 | 0.01 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr2_+_27010439 | 0.01 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.1 | 0.3 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.6 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.2 | GO:0003071 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.0 | 0.7 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |