Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
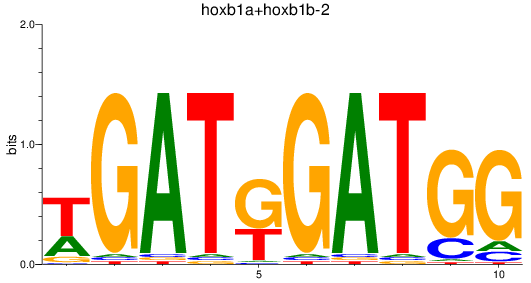
Results for hoxb1a+hoxb1b-2
Z-value: 0.11
Transcription factors associated with hoxb1a+hoxb1b-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb1a
|
ENSDARG00000008174 | homeobox B1a |
|
hoxb1b-2
|
ENSDARG00000112663 | homeobox B1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxb1a | dr11_v1_chr3_+_23768898_23768898 | -0.06 | 8.1e-01 | Click! |
Activity profile of hoxb1a+hoxb1b-2 motif
Sorted Z-values of hoxb1a+hoxb1b-2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_53143667 | 0.30 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr19_-_867071 | 0.27 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr2_-_49031303 | 0.20 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr17_+_19626479 | 0.20 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr4_-_10826575 | 0.19 |
ENSDART00000164771
ENSDART00000067256 |
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr6_-_4214297 | 0.17 |
ENSDART00000191433
|
trak2
|
trafficking protein, kinesin binding 2 |
| chr3_-_6417328 | 0.17 |
ENSDART00000160979
|
jpt1b
|
Jupiter microtubule associated homolog 1b |
| chr3_+_40409100 | 0.15 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr6_+_4255319 | 0.15 |
ENSDART00000170351
|
nbeal1
|
neurobeachin-like 1 |
| chr18_-_40753583 | 0.15 |
ENSDART00000026767
|
akt2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_+_28102487 | 0.14 |
ENSDART00000131638
ENSDART00000076265 |
zgc:91908
|
zgc:91908 |
| chr22_+_10676981 | 0.14 |
ENSDART00000138016
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr3_-_55650771 | 0.14 |
ENSDART00000162413
|
axin2
|
axin 2 (conductin, axil) |
| chr13_-_22961605 | 0.13 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr3_-_1283247 | 0.13 |
ENSDART00000149814
|
tcf20
|
transcription factor 20 |
| chr10_+_37182626 | 0.12 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr9_+_21795917 | 0.12 |
ENSDART00000169069
|
rev1
|
REV1, polymerase (DNA directed) |
| chr2_+_25658112 | 0.12 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr16_+_46401756 | 0.12 |
ENSDART00000147370
ENSDART00000144000 |
rpz2
|
rapunzel 2 |
| chr15_-_33304133 | 0.11 |
ENSDART00000186092
|
nbeab
|
neurobeachin b |
| chr2_+_25657958 | 0.11 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr8_-_49207319 | 0.10 |
ENSDART00000022870
|
fam110a
|
family with sequence similarity 110, member A |
| chr15_-_25094026 | 0.10 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr4_+_26496489 | 0.10 |
ENSDART00000160652
|
iqsec3a
|
IQ motif and Sec7 domain 3a |
| chr1_-_681116 | 0.10 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr3_-_55650417 | 0.10 |
ENSDART00000171441
|
axin2
|
axin 2 (conductin, axil) |
| chr23_+_27782071 | 0.10 |
ENSDART00000131379
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr15_-_25093680 | 0.09 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr5_+_31959954 | 0.09 |
ENSDART00000142826
|
myo1hb
|
myosin IHb |
| chr25_-_29072162 | 0.09 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr13_+_50778187 | 0.08 |
ENSDART00000159597
|
dntt
|
deoxynucleotidyltransferase, terminal |
| chr24_+_39518774 | 0.08 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr14_-_24101897 | 0.08 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr16_-_20932896 | 0.08 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr2_+_23731194 | 0.08 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr5_-_16791223 | 0.08 |
ENSDART00000192744
|
atad1a
|
ATPase family, AAA domain containing 1a |
| chr21_-_25395223 | 0.07 |
ENSDART00000016219
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr11_-_27702778 | 0.07 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr4_-_13255700 | 0.07 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr10_+_32646402 | 0.07 |
ENSDART00000137244
|
zbtb21
|
zinc finger and BTB domain containing 21 |
| chr11_-_40457325 | 0.06 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr14_-_7888748 | 0.06 |
ENSDART00000166293
|
ppp3cb
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_8964926 | 0.06 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr9_+_35017702 | 0.06 |
ENSDART00000193640
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr3_-_21118969 | 0.05 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr8_-_38201415 | 0.05 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr3_-_21062706 | 0.05 |
ENSDART00000155605
ENSDART00000153686 ENSDART00000157168 ENSDART00000156614 ENSDART00000155743 ENSDART00000156275 |
fam57ba
|
family with sequence similarity 57, member Ba |
| chr7_+_21787507 | 0.05 |
ENSDART00000100936
|
tmem88b
|
transmembrane protein 88 b |
| chr19_+_13994563 | 0.05 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr5_+_36896933 | 0.05 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr12_+_22580579 | 0.05 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr24_+_32472155 | 0.05 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr1_+_26626824 | 0.04 |
ENSDART00000158193
|
coro2a
|
coronin, actin binding protein, 2A |
| chr8_-_5220125 | 0.04 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr18_-_18875308 | 0.04 |
ENSDART00000127182
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr5_-_37881345 | 0.04 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr24_+_27548474 | 0.04 |
ENSDART00000105774
ENSDART00000123368 |
ek1
|
eph-like kinase 1 |
| chr5_+_6617401 | 0.04 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr15_-_46673161 | 0.04 |
ENSDART00000125211
|
CABZ01044053.1
|
|
| chr7_-_33350082 | 0.03 |
ENSDART00000008785
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr7_+_36898850 | 0.03 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr10_-_2527342 | 0.03 |
ENSDART00000184168
|
CU856539.1
|
|
| chr2_+_10821579 | 0.03 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr4_+_21129752 | 0.03 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr5_-_52277643 | 0.03 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr5_+_35398745 | 0.02 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr18_-_18874921 | 0.02 |
ENSDART00000193332
|
arl2bp
|
ADP-ribosylation factor-like 2 binding protein |
| chr17_-_20897407 | 0.02 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr11_+_30817943 | 0.02 |
ENSDART00000150130
ENSDART00000159997 |
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr3_+_29714775 | 0.02 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr7_-_37547889 | 0.02 |
ENSDART00000193359
|
cylda
|
cylindromatosis (turban tumor syndrome), a |
| chr6_+_19383267 | 0.02 |
ENSDART00000166549
|
mchr1a
|
melanin-concentrating hormone receptor 1a |
| chr17_-_28101880 | 0.02 |
ENSDART00000149458
ENSDART00000149226 ENSDART00000085758 |
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr2_+_10821127 | 0.02 |
ENSDART00000145770
ENSDART00000174629 ENSDART00000081094 |
glmna
|
glomulin, FKBP associated protein a |
| chr17_-_20897250 | 0.02 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr9_+_20781047 | 0.02 |
ENSDART00000139174
|
fam46c
|
family with sequence similarity 46, member C |
| chr18_+_28102620 | 0.01 |
ENSDART00000132342
|
kiaa1549lb
|
KIAA1549-like b |
| chr16_+_25196572 | 0.01 |
ENSDART00000141956
|
tyrobp
|
TYRO protein tyrosine kinase binding protein |
| chr20_+_1088658 | 0.01 |
ENSDART00000162991
|
BX537249.1
|
|
| chr7_+_7151832 | 0.01 |
ENSDART00000109485
|
gal3st3
|
galactose-3-O-sulfotransferase 3 |
| chr9_+_20780813 | 0.01 |
ENSDART00000142787
|
fam46c
|
family with sequence similarity 46, member C |
| chr25_+_2668892 | 0.01 |
ENSDART00000122929
|
bbs4
|
Bardet-Biedl syndrome 4 |
| chr9_-_53537989 | 0.01 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr10_-_25591194 | 0.01 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr8_-_39739056 | 0.01 |
ENSDART00000147992
|
si:ch211-170d8.5
|
si:ch211-170d8.5 |
| chr1_+_58370526 | 0.01 |
ENSDART00000067775
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr1_-_10577945 | 0.01 |
ENSDART00000179237
ENSDART00000040502 ENSDART00000186876 |
trpc5a
|
transient receptor potential cation channel, subfamily C, member 5a |
| chr9_+_22003942 | 0.01 |
ENSDART00000091013
|
si:dkey-57a22.15
|
si:dkey-57a22.15 |
| chr2_+_17451656 | 0.01 |
ENSDART00000163620
|
BX640408.1
|
|
| chr24_+_28528000 | 0.00 |
ENSDART00000155924
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr10_-_31175744 | 0.00 |
ENSDART00000191728
|
pknox2
|
pbx/knotted 1 homeobox 2 |
| chr8_-_39903803 | 0.00 |
ENSDART00000012391
|
cabp1a
|
calcium binding protein 1a |
| chr1_-_49505449 | 0.00 |
ENSDART00000187294
ENSDART00000132171 |
si:dkeyp-80c12.5
|
si:dkeyp-80c12.5 |
| chr8_+_6954984 | 0.00 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr8_+_6410933 | 0.00 |
ENSDART00000168789
|
lrrtm4l2
|
leucine rich repeat transmembrane neuronal 4 like 2 |
| chr2_+_22765492 | 0.00 |
ENSDART00000133222
|
espnlb
|
espin like b |
| chr1_-_9227804 | 0.00 |
ENSDART00000190360
|
gng13a
|
guanine nucleotide binding protein (G protein), gamma 13a |
| chr10_-_27049170 | 0.00 |
ENSDART00000143451
|
cnih2
|
cornichon family AMPA receptor auxiliary protein 2 |
| chr2_+_55984788 | 0.00 |
ENSDART00000183599
|
nmrk2
|
nicotinamide riboside kinase 2 |
| chr16_-_16761164 | 0.00 |
ENSDART00000135872
|
si:dkey-27n14.1
|
si:dkey-27n14.1 |
| chr24_-_21713883 | 0.00 |
ENSDART00000081357
|
flt3
|
fms-related tyrosine kinase 3 |
| chr10_-_44305180 | 0.00 |
ENSDART00000187552
|
CDK2AP1
|
cyclin dependent kinase 2 associated protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb1a+hoxb1b-2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.0 | 0.2 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.3 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |