Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
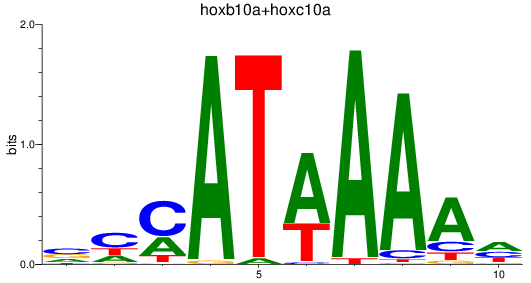
Results for hoxb10a+hoxc10a
Z-value: 0.25
Transcription factors associated with hoxb10a+hoxc10a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxb10a
|
ENSDARG00000011579 | homeobox B10a |
|
hoxc10a
|
ENSDARG00000070348 | homeobox C10a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxc10a | dr11_v1_chr23_+_35969228_35969228 | 0.93 | 3.6e-08 | Click! |
| hoxb10a | dr11_v1_chr3_+_23669267_23669267 | 0.84 | 1.2e-05 | Click! |
Activity profile of hoxb10a+hoxc10a motif
Sorted Z-values of hoxb10a+hoxc10a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_+_39606108 | 1.24 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr10_+_506538 | 1.14 |
ENSDART00000141713
|
si:ch211-242f23.3
|
si:ch211-242f23.3 |
| chr12_+_19356623 | 0.64 |
ENSDART00000078284
|
dmc1
|
DNA meiotic recombinase 1 |
| chr10_+_2587234 | 0.59 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr23_+_24085531 | 0.53 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr21_-_25756119 | 0.51 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr10_-_22845485 | 0.45 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr6_-_19683406 | 0.44 |
ENSDART00000158041
|
cfap52
|
cilia and flagella associated protein 52 |
| chr5_-_24174956 | 0.42 |
ENSDART00000137631
|
si:ch211-137i24.10
|
si:ch211-137i24.10 |
| chr21_-_13225402 | 0.41 |
ENSDART00000080347
|
wdr34
|
WD repeat domain 34 |
| chr13_-_37519774 | 0.41 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr6_+_30703828 | 0.37 |
ENSDART00000104628
|
insl5a
|
insulin-like 5a |
| chr5_+_30588856 | 0.37 |
ENSDART00000086661
|
dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr19_-_10881141 | 0.36 |
ENSDART00000162793
|
psmd4a
|
proteasome 26S subunit, non-ATPase 4a |
| chr5_-_30588539 | 0.35 |
ENSDART00000086686
ENSDART00000159163 ENSDART00000142958 |
c2cd2l
|
c2cd2-like |
| chr20_+_26538137 | 0.34 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr20_+_52546186 | 0.33 |
ENSDART00000110777
ENSDART00000153377 ENSDART00000153013 ENSDART00000042704 |
eef1db
|
eukaryotic translation elongation factor 1 delta b (guanine nucleotide exchange protein) |
| chr16_-_45917322 | 0.33 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr25_-_32449235 | 0.33 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr6_-_37468971 | 0.32 |
ENSDART00000126379
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr12_-_20373058 | 0.32 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr19_-_10881486 | 0.32 |
ENSDART00000168852
ENSDART00000160438 |
PSMD4 (1 of many)
psmd4a
|
proteasome 26S subunit, non-ATPase 4 proteasome 26S subunit, non-ATPase 4a |
| chr22_+_18187857 | 0.31 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr19_+_17385561 | 0.31 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr23_+_7379728 | 0.31 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr11_-_6265574 | 0.29 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr16_-_2558653 | 0.29 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr1_-_37377509 | 0.29 |
ENSDART00000113542
|
tnip2
|
TNFAIP3 interacting protein 2 |
| chr15_+_37559570 | 0.27 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr13_-_12660318 | 0.27 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr18_+_20825375 | 0.27 |
ENSDART00000090100
ENSDART00000146751 ENSDART00000141686 |
ttc23
|
tetratricopeptide repeat domain 23 |
| chr19_-_20163197 | 0.26 |
ENSDART00000148179
ENSDART00000151646 |
fam221a
|
family with sequence similarity 221, member A |
| chr22_+_18188045 | 0.26 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr7_-_26457208 | 0.25 |
ENSDART00000173519
|
zgc:172079
|
zgc:172079 |
| chr6_+_49551614 | 0.25 |
ENSDART00000022581
|
rab22a
|
RAB22A, member RAS oncogene family |
| chr7_+_22688781 | 0.24 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr5_-_26181863 | 0.23 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_+_26913120 | 0.23 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr16_-_21668082 | 0.23 |
ENSDART00000088513
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chr20_-_25533739 | 0.22 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr12_+_27117609 | 0.22 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr11_+_7580079 | 0.22 |
ENSDART00000091550
ENSDART00000193223 ENSDART00000193386 |
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr20_+_16173618 | 0.22 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr8_+_8860902 | 0.21 |
ENSDART00000144986
|
otud5a
|
OTU deubiquitinase 5a |
| chr13_-_50108337 | 0.21 |
ENSDART00000133308
|
nid1a
|
nidogen 1a |
| chr4_+_2267641 | 0.20 |
ENSDART00000165503
|
si:ch73-89b15.3
|
si:ch73-89b15.3 |
| chr10_-_34772211 | 0.20 |
ENSDART00000145450
ENSDART00000134307 |
dclk1a
|
doublecortin-like kinase 1a |
| chr6_-_39653972 | 0.20 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr24_-_27461295 | 0.20 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr24_-_12689571 | 0.19 |
ENSDART00000015517
|
pdcd6
|
programmed cell death 6 |
| chr8_+_2487883 | 0.19 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr10_+_40583930 | 0.19 |
ENSDART00000134523
|
taar18j
|
trace amine associated receptor 18j |
| chr5_+_36611128 | 0.19 |
ENSDART00000097684
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr17_+_33453689 | 0.19 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr24_+_7495945 | 0.18 |
ENSDART00000133525
ENSDART00000182460 ENSDART00000162954 |
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr13_+_22295905 | 0.18 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr1_-_35924495 | 0.18 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr25_+_18436301 | 0.18 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr19_+_7759354 | 0.17 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr8_+_18588551 | 0.17 |
ENSDART00000177476
ENSDART00000063539 |
prrg1
|
proline rich Gla (G-carboxyglutamic acid) 1 |
| chr2_+_2223837 | 0.17 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr22_-_4644484 | 0.17 |
ENSDART00000167748
|
fbn2b
|
fibrillin 2b |
| chr7_+_62248514 | 0.17 |
ENSDART00000025308
|
tbc1d19
|
TBC1 domain family, member 19 |
| chr6_+_35362225 | 0.17 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr7_+_25059845 | 0.17 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr19_+_5418006 | 0.16 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr16_+_25664078 | 0.16 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr19_+_4916233 | 0.16 |
ENSDART00000159512
|
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr16_+_25664387 | 0.16 |
ENSDART00000184394
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr4_+_7391110 | 0.15 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr24_-_31846366 | 0.15 |
ENSDART00000155295
|
steap2
|
STEAP family member 2, metalloreductase |
| chr10_+_26612321 | 0.15 |
ENSDART00000134322
|
fhl1b
|
four and a half LIM domains 1b |
| chr25_-_31423493 | 0.15 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr18_+_2189211 | 0.15 |
ENSDART00000170827
|
ccpg1
|
cell cycle progression 1 |
| chr5_-_48307804 | 0.15 |
ENSDART00000182831
ENSDART00000186920 ENSDART00000183585 |
mef2cb
|
myocyte enhancer factor 2cb |
| chr6_+_41191482 | 0.15 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr5_+_24245682 | 0.14 |
ENSDART00000049003
|
atp6v1aa
|
ATPase H+ transporting V1 subunit Aa |
| chr3_-_31804481 | 0.14 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr16_+_50969248 | 0.14 |
ENSDART00000172068
|
si:dkeyp-97a10.2
|
si:dkeyp-97a10.2 |
| chr18_-_20532339 | 0.13 |
ENSDART00000060291
|
ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr10_-_40589641 | 0.13 |
ENSDART00000140228
ENSDART00000154850 |
taar16b
|
trace amine associated receptor 16b |
| chr4_-_56898328 | 0.12 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr2_+_52847049 | 0.12 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr24_+_1042594 | 0.12 |
ENSDART00000109117
|
si:dkey-192l18.9
|
si:dkey-192l18.9 |
| chr10_-_2713228 | 0.12 |
ENSDART00000123754
ENSDART00000126236 |
mier3a
|
mesoderm induction early response 1, family member 3 a |
| chr19_+_19599528 | 0.12 |
ENSDART00000163054
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr23_+_10146542 | 0.11 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr10_-_29903165 | 0.11 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr2_-_55298075 | 0.11 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr23_+_27068225 | 0.11 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr18_+_19974289 | 0.11 |
ENSDART00000090334
ENSDART00000192982 |
skor1b
|
SKI family transcriptional corepressor 1b |
| chr14_-_33613794 | 0.11 |
ENSDART00000010022
|
xpnpep2
|
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr6_+_27624023 | 0.10 |
ENSDART00000147789
|
slco2a1
|
solute carrier organic anion transporter family, member 2A1 |
| chr5_+_15202495 | 0.10 |
ENSDART00000144915
|
tbx1
|
T-box 1 |
| chr16_-_7443388 | 0.10 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr6_+_36877968 | 0.10 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr20_+_27464721 | 0.10 |
ENSDART00000189552
|
kif26aa
|
kinesin family member 26Aa |
| chr17_+_33226955 | 0.10 |
ENSDART00000063333
|
pomca
|
proopiomelanocortin a |
| chr20_+_6543674 | 0.09 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr13_+_24584401 | 0.09 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr20_-_33961697 | 0.09 |
ENSDART00000061765
|
selp
|
selectin P |
| chr5_-_42883761 | 0.09 |
ENSDART00000167374
|
BX323596.2
|
|
| chr1_-_38195012 | 0.09 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr16_+_46459680 | 0.08 |
ENSDART00000101698
|
rpz3
|
rapunzel 3 |
| chr25_-_13614863 | 0.08 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr11_-_2700397 | 0.08 |
ENSDART00000082511
|
si:ch211-160o17.6
|
si:ch211-160o17.6 |
| chr22_+_25331766 | 0.08 |
ENSDART00000155006
ENSDART00000155836 |
si:ch211-226h8.15
|
si:ch211-226h8.15 |
| chr7_-_41338923 | 0.08 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr7_+_48761875 | 0.07 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr14_-_30967284 | 0.07 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr12_+_5081759 | 0.07 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr23_+_25354856 | 0.07 |
ENSDART00000109023
ENSDART00000147440 |
fmnl3
|
formin-like 3 |
| chr25_-_13614702 | 0.07 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr12_-_19091214 | 0.07 |
ENSDART00000153225
|
si:ch73-139e5.4
|
si:ch73-139e5.4 |
| chr19_+_2590182 | 0.07 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr7_+_27317174 | 0.07 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_+_5927255 | 0.06 |
ENSDART00000152866
|
si:ch211-168b3.2
|
si:ch211-168b3.2 |
| chr15_-_4580763 | 0.06 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr10_+_36650222 | 0.06 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr3_+_24134418 | 0.06 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr2_+_35595454 | 0.06 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr5_+_45140914 | 0.06 |
ENSDART00000172702
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_19191787 | 0.06 |
ENSDART00000152892
|
slc16a8
|
solute carrier family 16 (monocarboxylate transporter), member 8 |
| chr4_-_14192254 | 0.06 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr4_+_7391400 | 0.06 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr7_+_24165371 | 0.06 |
ENSDART00000173472
|
si:ch211-216p19.5
|
si:ch211-216p19.5 |
| chr18_+_7363242 | 0.05 |
ENSDART00000133375
|
si:dkey-30c15.17
|
si:dkey-30c15.17 |
| chr10_+_40606084 | 0.05 |
ENSDART00000133119
|
si:ch211-238p8.24
|
si:ch211-238p8.24 |
| chr2_+_20406399 | 0.05 |
ENSDART00000006817
ENSDART00000137848 |
palmda
|
palmdelphin a |
| chr21_+_4957685 | 0.05 |
ENSDART00000144166
|
B3GNT10
|
si:ch73-29c22.2 |
| chr8_+_2487250 | 0.05 |
ENSDART00000081325
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr14_-_31488100 | 0.05 |
ENSDART00000186246
|
cab39l1
|
calcium binding protein 39, like 1 |
| chr21_-_26520629 | 0.05 |
ENSDART00000142731
|
rce1b
|
Ras converting CAAX endopeptidase 1b |
| chr2_-_6039757 | 0.05 |
ENSDART00000013079
|
scp2a
|
sterol carrier protein 2a |
| chr6_+_52790049 | 0.05 |
ENSDART00000002571
|
matn4
|
matrilin 4 |
| chr8_+_30709685 | 0.05 |
ENSDART00000133989
|
upb1
|
ureidopropionase, beta |
| chr22_+_1421212 | 0.05 |
ENSDART00000161813
|
zgc:101130
|
zgc:101130 |
| chr11_+_30244356 | 0.05 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr25_+_4750972 | 0.04 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr12_-_13337033 | 0.04 |
ENSDART00000105903
ENSDART00000139786 |
lsm5
|
LSM5 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr19_+_19762183 | 0.04 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr7_-_45076131 | 0.04 |
ENSDART00000110590
|
zgc:194678
|
zgc:194678 |
| chr21_+_20396858 | 0.04 |
ENSDART00000003299
ENSDART00000146615 |
zgc:103482
|
zgc:103482 |
| chr24_-_27452488 | 0.04 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr8_-_19487327 | 0.04 |
ENSDART00000111710
|
edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr7_-_30087048 | 0.04 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr21_+_10756154 | 0.03 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr20_-_27390258 | 0.03 |
ENSDART00000010584
|
prph2l
|
peripherin 2, like |
| chr7_-_60096318 | 0.03 |
ENSDART00000189125
|
BX511067.1
|
|
| chr25_-_31739309 | 0.03 |
ENSDART00000098896
|
acot19
|
acyl-CoA thioesterase 19 |
| chr4_+_37406676 | 0.03 |
ENSDART00000130981
|
si:ch73-134f24.1
|
si:ch73-134f24.1 |
| chr18_-_17513426 | 0.03 |
ENSDART00000146725
|
b3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr17_-_2690083 | 0.03 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr19_+_42469058 | 0.02 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr16_-_21038015 | 0.02 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr17_+_45395846 | 0.02 |
ENSDART00000058793
|
nenf
|
neudesin neurotrophic factor |
| chr23_-_24047054 | 0.02 |
ENSDART00000184308
ENSDART00000185902 |
CR925720.1
|
|
| chr13_+_22264914 | 0.02 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr9_-_1939232 | 0.02 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr4_+_5741733 | 0.02 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr22_+_1779401 | 0.01 |
ENSDART00000170126
|
znf1154
|
zinc finger protein 1154 |
| chr24_-_8732519 | 0.01 |
ENSDART00000082351
|
tfap2a
|
transcription factor AP-2 alpha |
| chr24_+_12913329 | 0.01 |
ENSDART00000141829
|
si:dkeyp-28d2.4
|
si:dkeyp-28d2.4 |
| chr7_-_32599669 | 0.01 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr14_+_23184517 | 0.01 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr8_-_25120231 | 0.01 |
ENSDART00000147308
|
amigo1
|
adhesion molecule with Ig-like domain 1 |
| chr5_+_23136544 | 0.01 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr1_+_1915967 | 0.01 |
ENSDART00000131463
|
si:ch211-132g1.1
|
si:ch211-132g1.1 |
| chr9_-_41784799 | 0.01 |
ENSDART00000144573
ENSDART00000112542 ENSDART00000190486 |
obsl1b
|
obscurin-like 1b |
| chr15_-_28860282 | 0.01 |
ENSDART00000156738
|
gpr4
|
G protein-coupled receptor 4 |
| chr6_-_12588044 | 0.01 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr4_-_14191717 | 0.00 |
ENSDART00000147928
|
pus7l
|
pseudouridylate synthase 7-like |
| chr17_-_12389259 | 0.00 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr4_-_14191434 | 0.00 |
ENSDART00000142374
ENSDART00000136730 |
pus7l
|
pseudouridylate synthase 7-like |
| chr19_-_17385548 | 0.00 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr7_-_16562200 | 0.00 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr2_+_21855036 | 0.00 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr12_+_34051848 | 0.00 |
ENSDART00000153276
|
cyth1b
|
cytohesin 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxb10a+hoxc10a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.3 | GO:0034138 | toll-like receptor 2 signaling pathway(GO:0034134) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 0.3 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.2 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.1 | 0.1 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.0 | 0.1 | GO:0043282 | extraocular skeletal muscle development(GO:0002074) pharyngeal muscle development(GO:0043282) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.0 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 0.1 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.1 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.0 | 0.2 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0006833 | water transport(GO:0006833) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.0 | 0.0 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.0 | 0.1 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.0 | 0.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |