Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
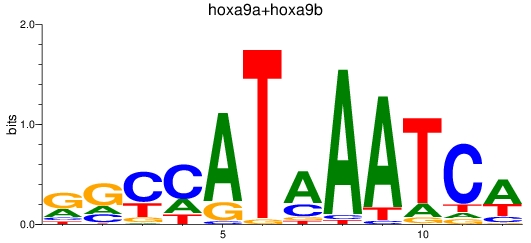
Results for hoxa9a+hoxa9b
Z-value: 0.94
Transcription factors associated with hoxa9a+hoxa9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa9b
|
ENSDARG00000056819 | homeobox A9b |
|
hoxa9a
|
ENSDARG00000105013 | homeobox A9a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa9b | dr11_v1_chr16_+_20915319_20915319 | -0.91 | 1.5e-07 | Click! |
| hoxa9a | dr11_v1_chr19_+_19747430_19747430 | -0.90 | 4.6e-07 | Click! |
Activity profile of hoxa9a+hoxa9b motif
Sorted Z-values of hoxa9a+hoxa9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_33647138 | 3.86 |
ENSDART00000142111
ENSDART00000015547 |
cldng
|
claudin g |
| chr17_-_4245902 | 3.00 |
ENSDART00000151851
|
gdf3
|
growth differentiation factor 3 |
| chr16_+_29509133 | 2.38 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr19_-_27550768 | 2.30 |
ENSDART00000142313
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr21_-_13690712 | 2.22 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr17_-_4245311 | 2.11 |
ENSDART00000055379
|
gdf3
|
growth differentiation factor 3 |
| chr17_-_2595736 | 2.10 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr7_+_30626378 | 2.04 |
ENSDART00000173533
ENSDART00000052541 |
ccnb2
|
cyclin B2 |
| chr10_-_8053753 | 2.04 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr19_-_31042570 | 1.96 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr6_-_42388608 | 1.94 |
ENSDART00000049425
|
sec61a1l
|
Sec61 translocon alpha 1 subunit, like |
| chr8_+_21229718 | 1.91 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr17_-_2590222 | 1.90 |
ENSDART00000185711
|
CR759892.1
|
|
| chr5_-_29514689 | 1.89 |
ENSDART00000126018
ENSDART00000125175 |
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr7_+_1473929 | 1.86 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr9_-_34937025 | 1.80 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr7_+_26545911 | 1.78 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr10_-_8046764 | 1.72 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr5_+_41476443 | 1.71 |
ENSDART00000145228
ENSDART00000137981 ENSDART00000142538 |
pias2
|
protein inhibitor of activated STAT, 2 |
| chr6_-_53144336 | 1.67 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr2_-_32738535 | 1.65 |
ENSDART00000135293
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr7_-_33684632 | 1.64 |
ENSDART00000130553
|
tle3b
|
transducin-like enhancer of split 3b |
| chr17_+_19626479 | 1.62 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr10_+_44699734 | 1.54 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr19_-_27830818 | 1.51 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr3_-_40232615 | 1.48 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr1_+_36771954 | 1.44 |
ENSDART00000149022
|
arhgap10
|
Rho GTPase activating protein 10 |
| chr1_+_31658011 | 1.36 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr7_+_51795667 | 1.35 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr8_-_2616326 | 1.35 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr20_-_49889111 | 1.33 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr8_+_3431671 | 1.30 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr4_-_4261673 | 1.26 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr22_-_20695237 | 1.26 |
ENSDART00000112722
|
org
|
oogenesis-related gene |
| chr3_+_58472305 | 1.26 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr10_+_44700103 | 1.26 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr22_-_11493236 | 1.26 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr8_+_15269423 | 1.25 |
ENSDART00000020386
|
gclm
|
glutamate-cysteine ligase, modifier subunit |
| chr7_+_30779761 | 1.25 |
ENSDART00000066806
ENSDART00000173671 |
mcee
|
methylmalonyl CoA epimerase |
| chr17_-_30652738 | 1.23 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr7_+_26545502 | 1.22 |
ENSDART00000140528
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr8_+_12951155 | 1.22 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr18_+_27515640 | 1.22 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr10_-_8053385 | 1.22 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr9_-_23807032 | 1.20 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr17_+_25833947 | 1.18 |
ENSDART00000044328
ENSDART00000154604 |
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr9_+_50175366 | 1.18 |
ENSDART00000170352
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr14_+_22132896 | 1.18 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr25_+_15997957 | 1.17 |
ENSDART00000140047
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr8_-_38355555 | 1.17 |
ENSDART00000134283
ENSDART00000132077 ENSDART00000146378 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr5_+_872299 | 1.16 |
ENSDART00000130042
|
fubp3
|
far upstream element (FUSE) binding protein 3 |
| chr8_-_38355268 | 1.15 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr22_-_10541712 | 1.15 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr6_+_38626926 | 1.14 |
ENSDART00000190339
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr2_+_22409249 | 1.10 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr9_+_492459 | 1.10 |
ENSDART00000112635
|
CU984600.1
|
|
| chr19_+_32321797 | 1.09 |
ENSDART00000167664
|
atxn1a
|
ataxin 1a |
| chr18_-_35407289 | 1.08 |
ENSDART00000012018
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr17_+_32622933 | 1.07 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr18_-_35407695 | 1.06 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr14_+_34490445 | 1.04 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr4_+_2482046 | 1.04 |
ENSDART00000103371
|
zdhhc17
|
zinc finger, DHHC-type containing 17 |
| chr18_-_35407530 | 1.04 |
ENSDART00000137663
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr24_-_21903588 | 1.03 |
ENSDART00000180991
|
spata13
|
spermatogenesis associated 13 |
| chr24_-_40901410 | 1.02 |
ENSDART00000170688
|
wdr48a
|
WD repeat domain 48a |
| chr5_-_36597612 | 1.02 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr11_-_35171768 | 1.02 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr9_-_30259295 | 1.01 |
ENSDART00000139106
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr9_-_30259447 | 1.00 |
ENSDART00000140585
ENSDART00000145052 ENSDART00000129926 |
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr18_+_11858397 | 1.00 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr22_-_10541372 | 1.00 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr20_-_16548912 | 0.98 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr19_+_27858866 | 0.98 |
ENSDART00000140336
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr8_-_31416403 | 0.98 |
ENSDART00000098912
|
znf131
|
zinc finger protein 131 |
| chr3_+_40255408 | 0.97 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr20_-_51814080 | 0.97 |
ENSDART00000041476
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr20_-_51831657 | 0.96 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr3_+_28502419 | 0.96 |
ENSDART00000151081
|
sept12
|
septin 12 |
| chr7_+_24881680 | 0.95 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr25_+_16880990 | 0.93 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr24_+_36317544 | 0.93 |
ENSDART00000048640
ENSDART00000156096 |
pus3
|
pseudouridylate synthase 3 |
| chr12_+_36428052 | 0.93 |
ENSDART00000131300
|
unk
|
unkempt family zinc finger |
| chr14_+_25505468 | 0.93 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr13_-_35907768 | 0.92 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr19_+_4968947 | 0.92 |
ENSDART00000003634
ENSDART00000134808 |
stard3
|
StAR-related lipid transfer (START) domain containing 3 |
| chr13_-_22961605 | 0.92 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr9_+_28232522 | 0.92 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr9_-_30258957 | 0.91 |
ENSDART00000144259
|
si:dkey-100n23.5
|
si:dkey-100n23.5 |
| chr20_-_51831816 | 0.91 |
ENSDART00000060505
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr17_+_26718648 | 0.90 |
ENSDART00000154492
|
nrde2
|
NRDE-2, necessary for RNA interference, domain containing |
| chr22_+_10713713 | 0.90 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr9_-_14137295 | 0.90 |
ENSDART00000127640
ENSDART00000189018 ENSDART00000188985 |
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr13_-_35908275 | 0.89 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr1_+_11107688 | 0.89 |
ENSDART00000109858
|
knstrn
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr9_-_12575776 | 0.88 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr2_+_34967022 | 0.87 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr25_+_16214854 | 0.87 |
ENSDART00000109672
ENSDART00000190093 |
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr8_-_30424182 | 0.87 |
ENSDART00000099021
|
dock8
|
dedicator of cytokinesis 8 |
| chr5_+_25733774 | 0.87 |
ENSDART00000137088
ENSDART00000098467 |
abhd17b
|
abhydrolase domain containing 17B |
| chr4_+_10754802 | 0.87 |
ENSDART00000181470
|
stab2
|
stabilin 2 |
| chr20_+_2642855 | 0.87 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr15_-_10341048 | 0.87 |
ENSDART00000171013
|
tenm4
|
teneurin transmembrane protein 4 |
| chr6_+_1724889 | 0.86 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr1_-_681116 | 0.86 |
ENSDART00000165894
|
adamts1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr10_-_1961930 | 0.85 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr23_-_3758637 | 0.85 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr12_+_22404108 | 0.84 |
ENSDART00000153055
|
hdlbpb
|
high density lipoprotein binding protein b |
| chr24_-_33801668 | 0.84 |
ENSDART00000079202
|
abcb8
|
ATP-binding cassette, sub-family B (MDR/TAP), member 8 |
| chr1_-_51038885 | 0.83 |
ENSDART00000035150
|
spast
|
spastin |
| chr23_+_43718115 | 0.81 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr19_-_30447611 | 0.81 |
ENSDART00000073705
ENSDART00000048977 ENSDART00000191237 |
abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr10_-_1961576 | 0.81 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr24_+_39211288 | 0.81 |
ENSDART00000061540
|
im:7160594
|
im:7160594 |
| chr22_+_21549419 | 0.81 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr7_+_59033902 | 0.81 |
ENSDART00000168888
|
fastkd3
|
FAST kinase domains 3 |
| chr23_-_3759345 | 0.80 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr1_+_12177195 | 0.80 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr16_+_55078477 | 0.79 |
ENSDART00000170074
|
azi2
|
5-azacytidine induced 2 |
| chr10_-_9961488 | 0.79 |
ENSDART00000191023
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr9_-_30502010 | 0.79 |
ENSDART00000149483
|
si:dkey-229b18.3
|
si:dkey-229b18.3 |
| chr14_-_24110062 | 0.78 |
ENSDART00000177062
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr19_-_82504 | 0.78 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr17_+_13031497 | 0.77 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr11_-_26832685 | 0.77 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr5_-_8682590 | 0.77 |
ENSDART00000142762
|
zgc:153352
|
zgc:153352 |
| chr3_-_11922713 | 0.77 |
ENSDART00000150575
|
coro7
|
coronin 7 |
| chr18_-_13121983 | 0.77 |
ENSDART00000092648
|
rxylt1
|
ribitol xylosyltransferase 1 |
| chr5_-_66823750 | 0.76 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr6_+_20954400 | 0.76 |
ENSDART00000143248
ENSDART00000165806 |
stk11ip
|
serine/threonine kinase 11 interacting protein |
| chr13_+_13945218 | 0.75 |
ENSDART00000089501
ENSDART00000142997 |
eif2ak3
|
eukaryotic translation initiation factor 2-alpha kinase 3 |
| chr2_-_37277626 | 0.75 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr1_-_52201266 | 0.74 |
ENSDART00000143805
ENSDART00000023757 |
rab3da
|
RAB3D, member RAS oncogene family, a |
| chr5_-_41124241 | 0.74 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr7_-_19369002 | 0.74 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr17_-_51262430 | 0.74 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr20_-_20821783 | 0.73 |
ENSDART00000152577
ENSDART00000027603 ENSDART00000145601 |
ckbb
|
creatine kinase, brain b |
| chr4_+_13586689 | 0.73 |
ENSDART00000067161
ENSDART00000138201 |
tnpo3
|
transportin 3 |
| chr2_-_44777592 | 0.72 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr14_+_30795559 | 0.72 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr13_+_49727333 | 0.71 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr20_-_51186524 | 0.71 |
ENSDART00000027836
ENSDART00000114407 |
rbm25b
|
RNA binding motif protein 25b |
| chr2_+_59041081 | 0.71 |
ENSDART00000067736
|
stk11
|
serine/threonine kinase 11 |
| chr2_-_37797577 | 0.70 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr18_-_39288894 | 0.70 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr23_-_20051369 | 0.69 |
ENSDART00000049836
|
bgnb
|
biglycan b |
| chr6_+_12527725 | 0.69 |
ENSDART00000149328
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr8_-_32803227 | 0.69 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr23_+_39963599 | 0.68 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr11_-_22916641 | 0.67 |
ENSDART00000080201
ENSDART00000154813 |
mdm4
|
MDM4, p53 regulator |
| chr4_+_1761029 | 0.67 |
ENSDART00000026963
|
pmpcb
|
peptidase (mitochondrial processing) beta |
| chr16_+_32014552 | 0.67 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr17_-_25630822 | 0.66 |
ENSDART00000126201
ENSDART00000105503 ENSDART00000151878 |
rab3gap2
|
RAB3 GTPase activating protein subunit 2 (non-catalytic) |
| chr17_-_8592824 | 0.66 |
ENSDART00000127022
|
CU462878.1
|
|
| chr10_-_33251876 | 0.66 |
ENSDART00000184565
|
bcl7ba
|
BCL tumor suppressor 7Ba |
| chr9_-_51563575 | 0.66 |
ENSDART00000167034
ENSDART00000148918 |
tank
|
TRAF family member-associated NFKB activator |
| chr6_+_27992886 | 0.66 |
ENSDART00000160354
|
amotl2a
|
angiomotin like 2a |
| chr1_-_58900851 | 0.66 |
ENSDART00000183085
ENSDART00000188855 ENSDART00000182567 |
CABZ01084501.3
|
Danio rerio microfibril-associated glycoprotein 4-like (LOC100334800), transcript variant 2, mRNA. |
| chr2_+_21185903 | 0.65 |
ENSDART00000131369
|
si:dkey-29d8.3
|
si:dkey-29d8.3 |
| chr9_+_6009077 | 0.65 |
ENSDART00000057484
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr4_-_13255700 | 0.64 |
ENSDART00000162277
ENSDART00000026593 |
grip1
|
glutamate receptor interacting protein 1 |
| chr16_-_17586883 | 0.64 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr21_+_39100289 | 0.64 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr11_-_40457325 | 0.64 |
ENSDART00000128442
|
tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1B |
| chr22_-_14272699 | 0.64 |
ENSDART00000190121
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr22_+_22437561 | 0.64 |
ENSDART00000089622
|
kif14
|
kinesin family member 14 |
| chr1_+_35985813 | 0.64 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr10_+_39199547 | 0.63 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr15_-_1843831 | 0.63 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_-_49854432 | 0.62 |
ENSDART00000180731
|
blvra
|
biliverdin reductase A |
| chr12_-_22238004 | 0.62 |
ENSDART00000038310
|
ormdl3
|
ORMDL sphingolipid biosynthesis regulator 3 |
| chr8_-_38201415 | 0.61 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_-_31517300 | 0.60 |
ENSDART00000164799
|
slc6a6b
|
solute carrier family 6 (neurotransmitter transporter), member 6b |
| chr5_-_62940851 | 0.60 |
ENSDART00000137052
|
specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr17_+_25331576 | 0.60 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr18_+_22138924 | 0.59 |
ENSDART00000183961
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr11_-_13341051 | 0.59 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr10_+_22034477 | 0.58 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr17_+_50701748 | 0.58 |
ENSDART00000191938
ENSDART00000183220 ENSDART00000049464 |
fermt2
|
fermitin family member 2 |
| chr7_-_14446512 | 0.57 |
ENSDART00000041577
|
kif7
|
kinesin family member 7 |
| chr16_+_3185541 | 0.57 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr20_+_38458084 | 0.56 |
ENSDART00000020153
ENSDART00000135912 |
coq8a
|
coenzyme Q8A |
| chr11_-_40147032 | 0.56 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr21_-_9769500 | 0.56 |
ENSDART00000170710
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr7_+_29863548 | 0.56 |
ENSDART00000136555
ENSDART00000134068 |
tln2a
|
talin 2a |
| chr21_+_11749097 | 0.56 |
ENSDART00000102408
ENSDART00000102404 |
ell2
|
elongation factor, RNA polymerase II, 2 |
| chr11_+_45153104 | 0.56 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr15_+_24704798 | 0.55 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr23_-_43718067 | 0.55 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr6_-_7123210 | 0.55 |
ENSDART00000041304
|
atg3
|
autophagy related 3 |
| chr1_+_24557414 | 0.54 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr20_-_43811023 | 0.54 |
ENSDART00000153134
|
mark3b
|
MAP/microtubule affinity-regulating kinase 3b |
| chr9_+_25840720 | 0.54 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr2_+_2168547 | 0.54 |
ENSDART00000029347
|
higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr3_+_21200763 | 0.54 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr23_+_5631381 | 0.54 |
ENSDART00000149143
|
pkp1a
|
plakophilin 1a |
| chr13_-_479129 | 0.54 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr3_+_52545400 | 0.53 |
ENSDART00000184183
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr21_+_20903244 | 0.53 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr4_+_28374628 | 0.52 |
ENSDART00000076037
|
alg10
|
asparagine-linked glycosylation 10 |
| chr1_+_38142354 | 0.51 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa9a+hoxa9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.9 | 2.8 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.8 | 3.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.6 | 1.7 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.3 | 1.0 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.0 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.3 | 1.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.3 | 0.8 | GO:0034214 | protein hexamerization(GO:0034214) mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 2.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.3 | 1.9 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.2 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.9 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.2 | 1.3 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 1.0 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.2 | 2.0 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 1.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 0.9 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 0.6 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.2 | 0.5 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.2 | 0.7 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.9 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.2 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 1.0 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.2 | 0.7 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.2 | 0.8 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 0.6 | GO:0090153 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 1.7 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 1.8 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 0.9 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.9 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 2.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.1 | 0.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 2.1 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.5 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.8 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 0.7 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.4 | GO:1902024 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.8 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.3 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.3 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.1 | 0.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 0.6 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.5 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.9 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 0.5 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 0.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.1 | 0.7 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 | 0.5 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.1 | 0.4 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.6 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.7 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.1 | 0.9 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 3.2 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.8 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 1.7 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.7 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.2 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.3 | GO:1901909 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 1.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.8 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.9 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.9 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.6 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 2.6 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.6 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 2.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.4 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 2.3 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.2 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 0.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0045840 | positive regulation of mitotic nuclear division(GO:0045840) positive regulation of nuclear division(GO:0051785) |
| 0.0 | 0.9 | GO:0030301 | cholesterol transport(GO:0030301) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.7 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.6 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.9 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.1 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.0 | 0.1 | GO:0061641 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 1.5 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.2 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 1.6 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 2.0 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 2.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.1 | GO:0090280 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.6 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.3 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 1.3 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 1.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.9 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.0 | 0.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.3 | 1.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.2 | 1.6 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.2 | 0.7 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.2 | 0.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.2 | 0.6 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.6 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 3.2 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 5.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.9 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 2.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 2.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 0.9 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 3.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.3 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 2.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.7 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 5.0 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.2 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 4.9 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.2 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.9 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 3.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.6 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.6 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 1.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.4 | 3.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 0.9 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 1.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 0.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.2 | 1.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 2.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 0.8 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 0.5 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.2 | 2.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 5.0 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.2 | 0.5 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.1 | 0.9 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.9 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.7 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.9 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.3 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.1 | 0.5 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.7 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.1 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.5 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.9 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.2 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 0.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.8 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.9 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 3.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.8 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 4.8 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.7 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 2.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 5.7 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 3.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 2.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.6 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 3.5 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 1.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.0 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.4 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 0.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 2.2 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 5.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.3 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.2 | 1.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.8 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 2.0 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 2.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.9 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 1.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.7 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 1.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 3.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.4 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |