Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
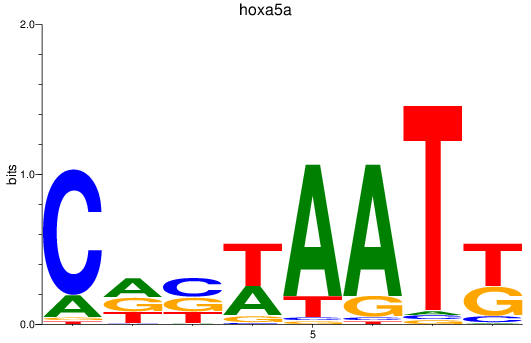
Results for hoxa5a
Z-value: 0.71
Transcription factors associated with hoxa5a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa5a
|
ENSDARG00000102501 | homeobox A5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa5a | dr11_v1_chr19_+_19759577_19759577 | 0.80 | 7.6e-05 | Click! |
Activity profile of hoxa5a motif
Sorted Z-values of hoxa5a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_50248152 | 2.56 |
ENSDART00000153998
|
zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr13_+_2908764 | 2.16 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr10_+_39091353 | 2.15 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr2_+_13462305 | 2.04 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr18_+_1154189 | 1.91 |
ENSDART00000135090
|
si:ch1073-75f15.2
|
si:ch1073-75f15.2 |
| chr16_-_25085327 | 1.72 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr11_+_25257022 | 1.66 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr23_-_42752550 | 1.65 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr15_-_28200049 | 1.59 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr16_-_41059919 | 1.50 |
ENSDART00000188803
|
si:dkey-201i6.8
|
si:dkey-201i6.8 |
| chr21_-_25618175 | 1.45 |
ENSDART00000133512
|
fosl1b
|
FOS-like antigen 1b |
| chr21_+_11885404 | 1.43 |
ENSDART00000092015
|
dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr10_+_26973063 | 1.29 |
ENSDART00000143162
ENSDART00000186210 |
tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr25_-_24233884 | 1.28 |
ENSDART00000146419
|
si:dkey-11e23.9
|
si:dkey-11e23.9 |
| chr25_-_13188214 | 1.22 |
ENSDART00000187298
|
si:ch211-147m6.1
|
si:ch211-147m6.1 |
| chr16_-_24195252 | 1.21 |
ENSDART00000136205
ENSDART00000048599 ENSDART00000161547 |
rps19
|
ribosomal protein S19 |
| chr2_-_43942595 | 1.20 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr11_-_30636163 | 1.20 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr8_+_18830759 | 1.20 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr3_-_59981476 | 1.19 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr4_-_11403811 | 1.17 |
ENSDART00000067272
ENSDART00000140018 |
pimr173
|
Pim proto-oncogene, serine/threonine kinase, related 173 |
| chr6_-_7720332 | 1.11 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr4_+_22480169 | 1.02 |
ENSDART00000146272
ENSDART00000066904 |
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr3_-_24456451 | 1.01 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr5_-_68022631 | 1.01 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr13_-_12006007 | 0.98 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr6_-_7718200 | 0.97 |
ENSDART00000189664
ENSDART00000114282 |
rpsa
|
ribosomal protein SA |
| chr19_-_41052408 | 0.96 |
ENSDART00000151120
|
sgce
|
sarcoglycan, epsilon |
| chr21_-_2952789 | 0.96 |
ENSDART00000182898
|
znf971
|
zinc finger protein 971 |
| chr16_-_12787029 | 0.90 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr16_-_35394422 | 0.89 |
ENSDART00000180593
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr13_-_23074238 | 0.86 |
ENSDART00000143210
|
srgn
|
serglycin |
| chr11_-_30634286 | 0.85 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr19_+_712127 | 0.83 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr2_+_31597841 | 0.79 |
ENSDART00000162181
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr2_-_20574193 | 0.79 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr15_-_47956388 | 0.79 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr22_-_18387059 | 0.76 |
ENSDART00000007769
|
tssk6
|
testis-specific serine kinase 6 |
| chr21_-_2299002 | 0.73 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr15_-_23692359 | 0.70 |
ENSDART00000141618
|
ercc2
|
excision repair cross-complementation group 2 |
| chr3_-_32898626 | 0.70 |
ENSDART00000103201
|
kat7a
|
K(lysine) acetyltransferase 7a |
| chr15_-_15469079 | 0.69 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr2_+_32826235 | 0.68 |
ENSDART00000143127
|
si:dkey-154p10.3
|
si:dkey-154p10.3 |
| chr3_-_30061985 | 0.68 |
ENSDART00000189583
|
ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
| chr5_-_26181863 | 0.68 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr11_-_30630628 | 0.68 |
ENSDART00000103265
|
zgc:158773
|
zgc:158773 |
| chr13_+_35635672 | 0.67 |
ENSDART00000148481
|
thbs2a
|
thrombospondin 2a |
| chr13_-_32626247 | 0.65 |
ENSDART00000100663
|
wdr35
|
WD repeat domain 35 |
| chr2_+_6127593 | 0.64 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr21_+_45510448 | 0.64 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr4_-_18851365 | 0.63 |
ENSDART00000021782
|
mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr13_-_12005429 | 0.62 |
ENSDART00000180302
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr11_-_8782871 | 0.62 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr1_-_11973341 | 0.60 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr7_-_50395395 | 0.59 |
ENSDART00000065868
|
vps33b
|
vacuolar protein sorting 33B |
| chr17_+_24615091 | 0.59 |
ENSDART00000064739
|
rpl13a
|
ribosomal protein L13a |
| chr5_-_20814576 | 0.58 |
ENSDART00000098682
ENSDART00000147639 |
si:ch211-225b11.1
|
si:ch211-225b11.1 |
| chr23_-_31506854 | 0.58 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr22_+_19366866 | 0.58 |
ENSDART00000137301
|
si:dkey-21e2.12
|
si:dkey-21e2.12 |
| chr2_+_38264964 | 0.55 |
ENSDART00000182068
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr13_+_27316934 | 0.54 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr23_-_46034609 | 0.53 |
ENSDART00000158712
|
zgc:65873
|
zgc:65873 |
| chr3_+_32112004 | 0.52 |
ENSDART00000105272
|
zgc:173593
|
zgc:173593 |
| chr23_+_4324625 | 0.50 |
ENSDART00000146302
ENSDART00000136792 ENSDART00000135027 ENSDART00000179819 |
sgk2a
|
serum/glucocorticoid regulated kinase 2a |
| chr13_-_8601184 | 0.50 |
ENSDART00000045086
|
prkceb
|
protein kinase C, epsilon b |
| chr7_-_24149670 | 0.49 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr22_+_37631234 | 0.48 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr25_-_16057539 | 0.47 |
ENSDART00000090388
|
cyb5r2
|
cytochrome b5 reductase 2 |
| chr6_-_47506351 | 0.46 |
ENSDART00000128700
|
im:7151449
|
im:7151449 |
| chr2_-_59145027 | 0.46 |
ENSDART00000128320
|
FO834803.1
|
|
| chr1_-_35924495 | 0.45 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr23_+_40109353 | 0.44 |
ENSDART00000149249
|
ghrhrl
|
growth hormone releasing hormone receptor, like |
| chr6_-_9236309 | 0.44 |
ENSDART00000160397
ENSDART00000164603 |
iqcb1
|
IQ motif containing B1 |
| chr7_+_34290051 | 0.44 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr10_+_36178713 | 0.44 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr3_-_62380146 | 0.44 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr20_+_52774730 | 0.42 |
ENSDART00000014606
|
phactr1
|
phosphatase and actin regulator 1 |
| chr17_-_43391499 | 0.42 |
ENSDART00000189280
|
itpk1b
|
inositol-tetrakisphosphate 1-kinase b |
| chr4_-_9852318 | 0.42 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr20_-_9095105 | 0.41 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr16_+_25664078 | 0.41 |
ENSDART00000154024
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr20_-_29483514 | 0.40 |
ENSDART00000062370
|
actc1a
|
actin, alpha, cardiac muscle 1a |
| chr21_-_4922981 | 0.40 |
ENSDART00000191819
ENSDART00000113556 ENSDART00000193269 ENSDART00000182961 |
kcnt1
|
potassium channel, subfamily T, member 1 |
| chr1_-_45614481 | 0.40 |
ENSDART00000148413
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr19_+_19600297 | 0.38 |
ENSDART00000160134
ENSDART00000183493 |
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr1_-_14332283 | 0.37 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr17_-_22324727 | 0.37 |
ENSDART00000160341
|
CU104709.1
|
|
| chr24_-_6078222 | 0.36 |
ENSDART00000146830
|
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr5_-_23909934 | 0.36 |
ENSDART00000142516
|
si:ch211-135f11.1
|
si:ch211-135f11.1 |
| chr22_-_36530902 | 0.36 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr16_+_25664387 | 0.35 |
ENSDART00000184394
|
nsmce2
|
NSE2/MMS21 homolog, SMC5-SMC6 complex SUMO ligase |
| chr18_+_11506561 | 0.32 |
ENSDART00000121647
|
PRMT8
|
protein arginine methyltransferase 8 |
| chr18_-_40901707 | 0.32 |
ENSDART00000139352
|
foxg1c
|
forkhead box G1c |
| chr11_-_42980535 | 0.32 |
ENSDART00000181160
ENSDART00000192064 |
CABZ01069998.1
|
|
| chr12_-_31461412 | 0.32 |
ENSDART00000175929
ENSDART00000186342 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr22_+_1462177 | 0.32 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr10_+_5025696 | 0.31 |
ENSDART00000031165
|
enoph1
|
enolase-phosphatase 1 |
| chr18_-_2433011 | 0.30 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr7_+_6480212 | 0.30 |
ENSDART00000173320
|
si:cabz01036022.1
|
si:cabz01036022.1 |
| chr13_+_27316632 | 0.30 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr14_+_6159162 | 0.30 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr5_+_66433287 | 0.29 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr5_-_16475682 | 0.29 |
ENSDART00000090695
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr4_-_10599062 | 0.27 |
ENSDART00000048003
|
tspan12
|
tetraspanin 12 |
| chr14_-_413273 | 0.27 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr8_-_18262149 | 0.27 |
ENSDART00000143000
|
rnf220b
|
ring finger protein 220b |
| chr15_+_23799461 | 0.25 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr8_+_20950401 | 0.25 |
ENSDART00000136484
ENSDART00000131329 ENSDART00000144451 ENSDART00000144929 |
si:dkeyp-82a1.1
|
si:dkeyp-82a1.1 |
| chr2_+_3533117 | 0.25 |
ENSDART00000132572
|
gpt2l
|
glutamic pyruvate transaminase (alanine aminotransferase) 2, like |
| chr22_+_17784414 | 0.25 |
ENSDART00000188189
ENSDART00000145260 |
tm6sf2
|
transmembrane 6 superfamily member 2 |
| chr5_-_25583125 | 0.25 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr7_+_22585447 | 0.24 |
ENSDART00000149144
|
chrnb1l
|
cholinergic receptor, nicotinic, beta 1 (muscle) like |
| chr25_-_20523927 | 0.24 |
ENSDART00000114448
|
si:ch211-269c21.2
|
si:ch211-269c21.2 |
| chr14_-_38826739 | 0.23 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr22_-_9896180 | 0.23 |
ENSDART00000138343
|
znf990
|
zinc finger protein 990 |
| chr4_-_30712588 | 0.23 |
ENSDART00000142393
|
si:dkey-16p19.5
|
si:dkey-16p19.5 |
| chr21_-_25668153 | 0.23 |
ENSDART00000192062
|
tmem179b
|
transmembrane protein 179B |
| chr24_-_21404367 | 0.23 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr10_+_21576909 | 0.22 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr15_-_42736433 | 0.22 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr7_+_42935126 | 0.22 |
ENSDART00000157747
|
BX284696.1
|
|
| chr5_-_67783593 | 0.21 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr14_-_30540967 | 0.21 |
ENSDART00000026907
|
her11
|
hairy-related 11 |
| chr19_+_19241372 | 0.21 |
ENSDART00000184392
ENSDART00000165008 |
ptpn23b
|
protein tyrosine phosphatase, non-receptor type 23, b |
| chr16_+_7242610 | 0.21 |
ENSDART00000081477
|
sri
|
sorcin |
| chr9_+_41080029 | 0.21 |
ENSDART00000141179
ENSDART00000019289 |
zgc:136439
|
zgc:136439 |
| chr11_-_38083397 | 0.20 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr5_+_59392183 | 0.19 |
ENSDART00000082983
ENSDART00000180882 |
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr4_+_11723852 | 0.19 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr7_-_6604623 | 0.19 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr7_+_23577723 | 0.18 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr2_-_49978227 | 0.18 |
ENSDART00000142835
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr8_-_4892913 | 0.18 |
ENSDART00000126808
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr23_+_5490854 | 0.18 |
ENSDART00000175403
|
tulp1a
|
tubby like protein 1a |
| chr2_+_37897079 | 0.17 |
ENSDART00000141784
|
tep1
|
telomerase-associated protein 1 |
| chr14_-_10762629 | 0.17 |
ENSDART00000061928
|
fgf16
|
fibroblast growth factor 16 |
| chr10_+_8690936 | 0.17 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr12_+_7406318 | 0.17 |
ENSDART00000152420
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr10_+_17776981 | 0.15 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr1_-_20273227 | 0.15 |
ENSDART00000146084
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr1_-_10647307 | 0.15 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr6_+_72040 | 0.14 |
ENSDART00000122957
|
crygm4
|
crystallin, gamma M4 |
| chr15_+_1372343 | 0.14 |
ENSDART00000152285
|
schip1
|
schwannomin interacting protein 1 |
| chr3_-_36750068 | 0.13 |
ENSDART00000173388
|
abcc6b.1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 6b, tandem duplicate 1 |
| chr22_+_1700552 | 0.13 |
ENSDART00000166185
|
znf1154
|
zinc finger protein 1154 |
| chr25_-_3627130 | 0.13 |
ENSDART00000171863
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr21_+_35327328 | 0.12 |
ENSDART00000130626
|
rars
|
arginyl-tRNA synthetase |
| chr8_-_30779101 | 0.12 |
ENSDART00000062229
|
p2rx7
|
purinergic receptor P2X, ligand-gated ion channel, 7 |
| chr4_-_65037243 | 0.12 |
ENSDART00000170059
|
si:ch211-283l16.1
|
si:ch211-283l16.1 |
| chr13_+_29771463 | 0.12 |
ENSDART00000134424
ENSDART00000138332 ENSDART00000134330 ENSDART00000160944 ENSDART00000076992 ENSDART00000160921 |
pax2a
|
paired box 2a |
| chr13_+_25422009 | 0.11 |
ENSDART00000057686
|
calhm2
|
calcium homeostasis modulator 2 |
| chr16_-_48074540 | 0.11 |
ENSDART00000187258
ENSDART00000186694 |
CSMD3 (1 of many)
|
si:ch211-236p22.1 |
| chr13_-_37383625 | 0.11 |
ENSDART00000192888
ENSDART00000100322 ENSDART00000138934 |
kcnh5b
|
potassium voltage-gated channel, subfamily H (eag-related), member 5b |
| chr8_-_22274222 | 0.11 |
ENSDART00000131805
|
nphp4
|
nephronophthisis 4 |
| chr3_+_17346502 | 0.11 |
ENSDART00000187786
ENSDART00000131584 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr4_-_14162327 | 0.10 |
ENSDART00000138656
|
si:dkey-234l24.1
|
si:dkey-234l24.1 |
| chr3_-_18410968 | 0.10 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr8_-_42594380 | 0.10 |
ENSDART00000140126
ENSDART00000135238 ENSDART00000192764 |
dok2
|
docking protein 2 |
| chr13_-_28688104 | 0.10 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr21_+_35327589 | 0.10 |
ENSDART00000145191
ENSDART00000132743 |
rars
|
arginyl-tRNA synthetase |
| chr3_+_32365811 | 0.09 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr11_+_10975481 | 0.09 |
ENSDART00000160488
|
itgb6
|
integrin, beta 6 |
| chr2_+_8779164 | 0.09 |
ENSDART00000134308
|
zzz3
|
zinc finger, ZZ-type containing 3 |
| chr10_+_16069987 | 0.08 |
ENSDART00000043936
|
megf10
|
multiple EGF-like-domains 10 |
| chr17_+_8542203 | 0.08 |
ENSDART00000158873
|
CU462878.2
|
|
| chr8_-_50287949 | 0.08 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr1_-_11850353 | 0.08 |
ENSDART00000033361
|
ttyh3b
|
tweety family member 3b |
| chr3_-_9488350 | 0.07 |
ENSDART00000186707
|
FO904885.2
|
|
| chr8_+_43053519 | 0.07 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr18_+_38321039 | 0.07 |
ENSDART00000132534
ENSDART00000111260 ENSDART00000192806 |
alx4b
|
ALX homeobox 4b |
| chr3_-_55404985 | 0.07 |
ENSDART00000154274
|
mafga
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Ga |
| chr12_+_28574863 | 0.07 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr7_-_47850702 | 0.07 |
ENSDART00000109511
|
si:ch211-186j3.6
|
si:ch211-186j3.6 |
| chr21_+_5580948 | 0.07 |
ENSDART00000160373
|
ly6m7
|
lymphocyte antigen 6 family member M7 |
| chr9_+_52492639 | 0.06 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr15_-_15230264 | 0.06 |
ENSDART00000155400
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr16_-_23379464 | 0.06 |
ENSDART00000045891
|
trim46a
|
tripartite motif containing 46a |
| chr19_+_21919856 | 0.05 |
ENSDART00000187306
ENSDART00000138544 |
galr1a
|
galanin receptor 1a |
| chr21_+_11509461 | 0.05 |
ENSDART00000134620
|
entpd2b
|
ectonucleoside triphosphate diphosphohydrolase 2b |
| chr18_+_13300768 | 0.05 |
ENSDART00000137506
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr14_-_2322484 | 0.05 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr16_-_11962959 | 0.04 |
ENSDART00000135379
|
cd4-2.2
|
CD4-2 molecule, tandem duplicate 2 |
| chr9_-_390874 | 0.04 |
ENSDART00000165324
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr5_+_38913621 | 0.04 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr7_-_33961551 | 0.04 |
ENSDART00000100104
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr7_-_69983462 | 0.04 |
ENSDART00000123380
|
KCNIP4
|
potassium voltage-gated channel interacting protein 4 |
| chr4_-_14173138 | 0.03 |
ENSDART00000142487
|
si:dkey-234l24.7
|
si:dkey-234l24.7 |
| chr1_-_9940494 | 0.03 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr17_-_43558494 | 0.02 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr7_-_41964877 | 0.02 |
ENSDART00000092351
ENSDART00000193395 ENSDART00000187947 |
neto2b
|
neuropilin (NRP) and tolloid (TLL)-like 2b |
| chr7_+_2236317 | 0.01 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr6_-_39489190 | 0.01 |
ENSDART00000151299
|
scn8ab
|
sodium channel, voltage gated, type VIII, alpha subunit b |
| chr9_+_21843915 | 0.01 |
ENSDART00000101977
|
kctd12.1
|
potassium channel tetramerisation domain containing 12.1 |
| chr15_-_1745408 | 0.01 |
ENSDART00000182311
|
stx1a
|
syntaxin 1A (brain) |
| chr23_-_44965582 | 0.01 |
ENSDART00000163367
|
tfr2
|
transferrin receptor 2 |
| chr24_+_35981471 | 0.01 |
ENSDART00000157373
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr23_+_21414005 | 0.01 |
ENSDART00000144686
|
iffo2a
|
intermediate filament family orphan 2a |
| chr15_+_8335600 | 0.00 |
ENSDART00000190066
ENSDART00000152603 |
egln2
|
egl-9 family hypoxia-inducible factor 2 |
| chr4_+_57099307 | 0.00 |
ENSDART00000131654
|
si:ch211-238e22.2
|
si:ch211-238e22.2 |
| chr17_+_6828550 | 0.00 |
ENSDART00000153777
|
si:ch73-242m19.1
|
si:ch73-242m19.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa5a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0019677 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.6 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 | 3.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.4 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.6 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.3 | GO:2000191 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.7 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:0000966 | RNA 5'-end processing(GO:0000966) germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.4 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 1.0 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 1.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.4 | GO:0036372 | opsin transport(GO:0036372) |
| 0.1 | 0.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 1.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.2 | GO:2001279 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.8 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.0 | 0.1 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.8 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.7 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.4 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.8 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 3.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.3 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 0.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.8 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.1 | 0.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.6 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 2.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.5 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.1 | 1.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 1.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |