Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
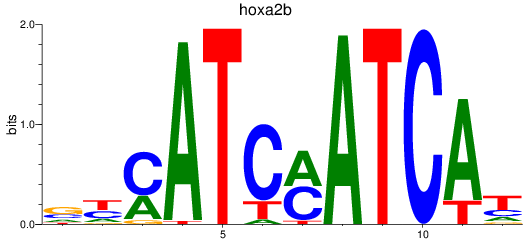
Results for hoxa2b
Z-value: 0.31
Transcription factors associated with hoxa2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
hoxa2b
|
ENSDARG00000023031 | homeobox A2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| hoxa2b | dr11_v1_chr16_+_20926673_20926673 | -0.43 | 7.8e-02 | Click! |
Activity profile of hoxa2b motif
Sorted Z-values of hoxa2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_4461104 | 0.79 |
ENSDART00000023090
ENSDART00000140770 |
slc12a10.1
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 10, tandem duplicate 1 |
| chr18_-_26715156 | 0.61 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr18_-_26715655 | 0.61 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr21_-_43656125 | 0.44 |
ENSDART00000136392
ENSDART00000192805 ENSDART00000181960 |
si:ch211-263m18.4
si:dkey-229d11.5
|
si:ch211-263m18.4 si:dkey-229d11.5 |
| chr9_-_712308 | 0.42 |
ENSDART00000144625
|
ndufb3
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3 |
| chr6_+_3640381 | 0.41 |
ENSDART00000172078
|
col28a2b
|
collagen, type XXVIII, alpha 2b |
| chr1_+_16144615 | 0.36 |
ENSDART00000054707
|
tusc3
|
tumor suppressor candidate 3 |
| chr24_+_5789790 | 0.35 |
ENSDART00000189600
|
BX470065.1
|
|
| chr1_+_24376394 | 0.33 |
ENSDART00000102542
|
zgc:171517
|
zgc:171517 |
| chr10_-_8053753 | 0.32 |
ENSDART00000162289
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_-_8046764 | 0.32 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr25_-_8660726 | 0.29 |
ENSDART00000168010
ENSDART00000171709 ENSDART00000163426 |
unc45a
|
unc-45 myosin chaperone A |
| chr1_-_56948694 | 0.28 |
ENSDART00000152597
|
si:ch211-1f22.1
|
si:ch211-1f22.1 |
| chr9_+_8365398 | 0.28 |
ENSDART00000138713
ENSDART00000136847 |
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr11_+_36243774 | 0.26 |
ENSDART00000023323
|
zgc:172270
|
zgc:172270 |
| chr24_+_5811808 | 0.25 |
ENSDART00000132428
|
si:ch211-157j23.3
|
si:ch211-157j23.3 |
| chr6_+_13201358 | 0.24 |
ENSDART00000190290
|
CT009620.1
|
|
| chr20_+_48129759 | 0.24 |
ENSDART00000148494
|
tram2
|
translocation associated membrane protein 2 |
| chr5_+_4298636 | 0.23 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr2_+_37295088 | 0.23 |
ENSDART00000056519
|
gpr160
|
G protein-coupled receptor 160 |
| chr24_-_6898302 | 0.23 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr24_+_5789582 | 0.23 |
ENSDART00000141504
|
BX470065.1
|
|
| chr9_+_8390737 | 0.22 |
ENSDART00000190891
ENSDART00000176877 ENSDART00000144851 ENSDART00000133880 ENSDART00000142233 |
zgc:113984
|
zgc:113984 |
| chr23_+_44611864 | 0.21 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr14_-_24101897 | 0.21 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr8_-_38355555 | 0.21 |
ENSDART00000134283
ENSDART00000132077 ENSDART00000146378 |
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr7_-_8324927 | 0.20 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr3_+_23752150 | 0.19 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr12_-_4301234 | 0.19 |
ENSDART00000152377
ENSDART00000152521 |
ca15b
|
carbonic anhydrase XVb |
| chr18_+_20532618 | 0.18 |
ENSDART00000006109
|
agk
|
acylglycerol kinase |
| chr3_+_18936009 | 0.18 |
ENSDART00000140330
|
si:ch211-198m1.1
|
si:ch211-198m1.1 |
| chr22_+_25088999 | 0.17 |
ENSDART00000158225
|
rrbp1b
|
ribosome binding protein 1b |
| chr7_-_59564011 | 0.17 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr2_-_7605121 | 0.17 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr6_-_55585423 | 0.17 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr8_+_23105117 | 0.17 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr21_-_32781612 | 0.17 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr18_+_17600570 | 0.17 |
ENSDART00000175258
ENSDART00000151850 ENSDART00000151934 |
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr20_-_438646 | 0.16 |
ENSDART00000009196
|
ufl1
|
UFM1-specific ligase 1 |
| chr15_+_20799943 | 0.16 |
ENSDART00000154665
|
aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr7_+_51774502 | 0.16 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr13_-_44423 | 0.16 |
ENSDART00000093247
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr10_-_45210184 | 0.16 |
ENSDART00000167128
|
pargl
|
poly (ADP-ribose) glycohydrolase, like |
| chr2_-_10821053 | 0.16 |
ENSDART00000056034
|
rpap2
|
RNA polymerase II associated protein 2 |
| chr8_+_36509885 | 0.15 |
ENSDART00000109530
|
slc7a4
|
solute carrier family 7, member 4 |
| chr24_+_7884880 | 0.15 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr22_+_38914983 | 0.15 |
ENSDART00000085701
|
senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr1_+_16127825 | 0.15 |
ENSDART00000122503
|
tusc3
|
tumor suppressor candidate 3 |
| chr9_-_27391908 | 0.15 |
ENSDART00000135221
|
nepro
|
nucleolus and neural progenitor protein |
| chr22_+_26290209 | 0.15 |
ENSDART00000060898
|
mrps28
|
mitochondrial ribosomal protein S28 |
| chr10_+_33627429 | 0.15 |
ENSDART00000138542
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr3_+_4502066 | 0.14 |
ENSDART00000088610
|
rangap1a
|
RAN GTPase activating protein 1a |
| chr5_-_32332560 | 0.14 |
ENSDART00000083862
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr4_+_77920666 | 0.14 |
ENSDART00000129523
|
ttll1
|
tubulin tyrosine ligase-like family, member 1 |
| chr17_-_49995366 | 0.14 |
ENSDART00000075223
|
cox7a2a
|
cytochrome c oxidase subunit VIIa polypeptide 2a (liver) |
| chr22_+_39084829 | 0.13 |
ENSDART00000002826
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr2_+_27855102 | 0.13 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr24_+_13735616 | 0.13 |
ENSDART00000184267
|
msc
|
musculin (activated B-cell factor-1) |
| chr17_+_34244345 | 0.13 |
ENSDART00000006058
|
eif2s1a
|
eukaryotic translation initiation factor 2, subunit 1 alpha a |
| chr13_+_17468161 | 0.13 |
ENSDART00000008906
|
znf503
|
zinc finger protein 503 |
| chr25_-_29072162 | 0.13 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr19_+_11978209 | 0.13 |
ENSDART00000111568
|
polr2k
|
polymerase (RNA) II (DNA directed) polypeptide K |
| chr8_-_38355268 | 0.13 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr5_+_69278089 | 0.13 |
ENSDART00000136656
|
selenom
|
selenoprotein M |
| chr2_+_27855346 | 0.13 |
ENSDART00000175159
ENSDART00000192645 |
buc
|
bucky ball |
| chr4_+_736395 | 0.13 |
ENSDART00000150698
|
ost4
|
oligosaccharyltransferase complex subunit 4 (non-catalytic) |
| chr3_+_58167288 | 0.13 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr10_+_7636811 | 0.13 |
ENSDART00000160673
|
hint1
|
histidine triad nucleotide binding protein 1 |
| chr20_+_18703108 | 0.13 |
ENSDART00000181188
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr24_+_20536056 | 0.13 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr19_+_1835940 | 0.13 |
ENSDART00000167372
|
ptk2aa
|
protein tyrosine kinase 2aa |
| chr22_-_10591876 | 0.13 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr4_+_74141400 | 0.13 |
ENSDART00000166994
|
trim24
|
tripartite motif containing 24 |
| chr20_-_20932760 | 0.12 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_-_39218609 | 0.12 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr3_-_34717882 | 0.12 |
ENSDART00000151127
|
thraa
|
thyroid hormone receptor alpha a |
| chr12_+_33975065 | 0.12 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr4_-_25485404 | 0.12 |
ENSDART00000041402
|
pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr16_+_3185541 | 0.12 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr10_-_33379850 | 0.12 |
ENSDART00000186924
|
ska2
|
spindle and kinetochore associated complex subunit 2 |
| chr21_-_23307653 | 0.12 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr20_+_18703695 | 0.12 |
ENSDART00000137766
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr24_+_34113424 | 0.12 |
ENSDART00000105572
|
gbx1
|
gastrulation brain homeobox 1 |
| chr13_-_44834895 | 0.12 |
ENSDART00000135573
|
si:dkeyp-2e4.5
|
si:dkeyp-2e4.5 |
| chr13_+_8604710 | 0.12 |
ENSDART00000091097
|
socs5b
|
suppressor of cytokine signaling 5b |
| chr22_-_26865361 | 0.12 |
ENSDART00000182504
|
hmox2a
|
heme oxygenase 2a |
| chr10_+_35417099 | 0.12 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr7_+_31145386 | 0.12 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr19_+_167612 | 0.11 |
ENSDART00000169574
|
tatdn1
|
TatD DNase domain containing 1 |
| chr14_+_48960078 | 0.11 |
ENSDART00000165514
|
mif
|
macrophage migration inhibitory factor |
| chr4_-_16824556 | 0.11 |
ENSDART00000165289
ENSDART00000185839 |
gys2
|
glycogen synthase 2 |
| chr1_+_41454210 | 0.11 |
ENSDART00000148251
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr16_+_12022543 | 0.11 |
ENSDART00000012673
|
gnb3a
|
guanine nucleotide binding protein (G protein), beta polypeptide 3a |
| chr9_+_50000504 | 0.11 |
ENSDART00000164409
ENSDART00000165451 ENSDART00000166509 |
slc38a11
|
solute carrier family 38, member 11 |
| chr7_+_38529263 | 0.11 |
ENSDART00000109495
ENSDART00000173804 |
nudt19
|
nudix (nucleoside diphosphate linked moiety X)-type motif 19 |
| chr3_-_59981476 | 0.11 |
ENSDART00000035878
ENSDART00000124038 |
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr19_+_28256076 | 0.11 |
ENSDART00000133354
|
irx4b
|
iroquois homeobox 4b |
| chr22_-_2959005 | 0.11 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr23_-_44819100 | 0.11 |
ENSDART00000076373
|
st8sia7.1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 7.1 |
| chr6_+_43015916 | 0.11 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr2_+_11028923 | 0.11 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr23_+_4689626 | 0.11 |
ENSDART00000131532
|
gp9
|
glycoprotein IX (platelet) |
| chr13_+_22480496 | 0.11 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr2_+_29249204 | 0.11 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr3_+_20001608 | 0.11 |
ENSDART00000137944
|
asb16
|
ankyrin repeat and SOCS box containing 16 |
| chr22_-_12726556 | 0.11 |
ENSDART00000105776
|
fam207a
|
family with sequence similarity 207, member A |
| chr8_+_27807266 | 0.11 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr3_+_32862730 | 0.10 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr8_+_27807974 | 0.10 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr1_-_9277986 | 0.10 |
ENSDART00000146065
ENSDART00000114876 ENSDART00000132812 |
ubn1
|
ubinuclein 1 |
| chr25_-_37314700 | 0.10 |
ENSDART00000017807
|
igl4v8
|
immunoglobulin light 4 variable 8 |
| chr22_+_14836040 | 0.10 |
ENSDART00000180951
|
gtpbp1l
|
GTP binding protein 1, like |
| chr7_-_40657831 | 0.10 |
ENSDART00000084153
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr3_-_29671653 | 0.10 |
ENSDART00000132083
|
sult3st1
|
sulfotransferase family 3, cytosolic sulfotransferase 1 |
| chr22_+_39084107 | 0.10 |
ENSDART00000191346
|
gmppb
|
GDP-mannose pyrophosphorylase B |
| chr6_-_13206255 | 0.10 |
ENSDART00000065373
|
eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_+_3501669 | 0.10 |
ENSDART00000160808
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr11_+_7276983 | 0.10 |
ENSDART00000172407
ENSDART00000161176 ENSDART00000161041 ENSDART00000165536 |
si:ch73-238c9.1
|
si:ch73-238c9.1 |
| chr17_-_49481672 | 0.10 |
ENSDART00000166394
|
fcf1
|
FCF1 rRNA-processing protein |
| chr18_+_3572314 | 0.10 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr4_-_16824231 | 0.10 |
ENSDART00000014007
|
gys2
|
glycogen synthase 2 |
| chr14_-_6666854 | 0.10 |
ENSDART00000133031
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr2_+_47623202 | 0.10 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr3_-_40528333 | 0.10 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr12_+_26491637 | 0.09 |
ENSDART00000087036
|
armc7
|
armadillo repeat containing 7 |
| chr21_+_25777425 | 0.09 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr23_+_36340520 | 0.09 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr12_+_33460794 | 0.09 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr9_-_40683722 | 0.09 |
ENSDART00000141979
ENSDART00000181228 |
bard1
|
BRCA1 associated RING domain 1 |
| chr10_-_7666810 | 0.09 |
ENSDART00000191479
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr1_-_44812014 | 0.09 |
ENSDART00000139044
|
si:dkey-9i23.8
|
si:dkey-9i23.8 |
| chr13_-_30034754 | 0.09 |
ENSDART00000143745
ENSDART00000041702 |
trmt2b
|
tRNA methyltransferase 2 homolog B |
| chr10_+_76864 | 0.09 |
ENSDART00000036375
|
vps26c
|
Down syndrome critical region 3 |
| chr24_-_6897884 | 0.09 |
ENSDART00000080766
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr11_-_25538341 | 0.09 |
ENSDART00000171560
|
si:dkey-245f22.3
|
si:dkey-245f22.3 |
| chr18_+_6126506 | 0.09 |
ENSDART00000125725
|
si:ch1073-390k14.1
|
si:ch1073-390k14.1 |
| chr3_-_61116258 | 0.09 |
ENSDART00000009194
ENSDART00000156978 |
aimp2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr7_-_41013575 | 0.09 |
ENSDART00000150139
|
insig1
|
insulin induced gene 1 |
| chr3_-_8510201 | 0.09 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr5_-_67292690 | 0.09 |
ENSDART00000062366
|
rsrc2
|
arginine/serine-rich coiled-coil 2 |
| chr12_+_1492641 | 0.09 |
ENSDART00000152411
|
usp22
|
ubiquitin specific peptidase 22 |
| chr14_+_3287740 | 0.09 |
ENSDART00000186290
|
cdx1a
|
caudal type homeobox 1a |
| chr24_-_31904924 | 0.09 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr16_-_2650341 | 0.09 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr14_-_23684814 | 0.09 |
ENSDART00000024604
|
mars2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr21_-_23308286 | 0.09 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr17_-_30839338 | 0.09 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr21_+_44581654 | 0.09 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr14_+_44774089 | 0.09 |
ENSDART00000098643
|
tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr12_+_2804505 | 0.08 |
ENSDART00000152193
|
mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr7_-_51793333 | 0.08 |
ENSDART00000180654
|
BX957362.5
|
|
| chr9_+_3548529 | 0.08 |
ENSDART00000130861
ENSDART00000123074 ENSDART00000147650 |
tlk1a
|
tousled-like kinase 1a |
| chr3_-_21118969 | 0.08 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr12_+_39685485 | 0.08 |
ENSDART00000163403
|
LO017650.1
|
|
| chr7_+_52154215 | 0.08 |
ENSDART00000098712
|
TMEM208
|
zgc:77041 |
| chr25_+_11456696 | 0.08 |
ENSDART00000171408
|
AGBL1
|
si:ch73-141f14.1 |
| chr3_-_32596394 | 0.08 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr4_+_77919987 | 0.08 |
ENSDART00000150171
|
ttll1
|
tubulin tyrosine ligase-like family, member 1 |
| chr1_-_43892349 | 0.08 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr8_-_38201415 | 0.08 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr1_-_43727418 | 0.08 |
ENSDART00000133715
ENSDART00000074597 ENSDART00000132542 ENSDART00000181792 |
bdh2
SLC9B2
|
3-hydroxybutyrate dehydrogenase, type 2 si:dkey-162b23.4 |
| chr14_+_109016 | 0.08 |
ENSDART00000158405
|
gpc2
|
glypican 2 |
| chr1_+_35985813 | 0.08 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr10_-_39011514 | 0.08 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr20_-_31496679 | 0.08 |
ENSDART00000153437
ENSDART00000153193 |
sash1a
|
SAM and SH3 domain containing 1a |
| chr20_+_18703454 | 0.08 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr22_-_382955 | 0.07 |
ENSDART00000082406
|
ora3
|
olfactory receptor class A related 3 |
| chr23_+_44881020 | 0.07 |
ENSDART00000149355
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr22_+_35089031 | 0.07 |
ENSDART00000076040
|
srfa
|
serum response factor a |
| chr9_+_2333550 | 0.07 |
ENSDART00000016417
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr7_-_35066457 | 0.07 |
ENSDART00000058067
|
zgc:112160
|
zgc:112160 |
| chr7_+_35075847 | 0.07 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr4_-_17822447 | 0.07 |
ENSDART00000185502
|
dnajb9b
|
DnaJ (Hsp40) homolog, subfamily B, member 9b |
| chr11_-_18017287 | 0.07 |
ENSDART00000155443
|
qrich1
|
glutamine-rich 1 |
| chr21_-_37075542 | 0.07 |
ENSDART00000129439
|
BX649250.1
|
|
| chr6_+_6802582 | 0.07 |
ENSDART00000189422
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr7_-_6431158 | 0.07 |
ENSDART00000173199
|
si:ch1073-153i20.5
|
si:ch1073-153i20.5 |
| chr16_-_50952266 | 0.07 |
ENSDART00000165408
|
si:dkeyp-97a10.3
|
si:dkeyp-97a10.3 |
| chr2_-_26001173 | 0.07 |
ENSDART00000010615
|
cldn11a
|
claudin 11a |
| chr5_-_19006290 | 0.07 |
ENSDART00000137022
|
golga3
|
golgin A3 |
| chr21_-_44081540 | 0.07 |
ENSDART00000130833
|
FO704810.1
|
|
| chr20_-_8443425 | 0.07 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_+_5875268 | 0.07 |
ENSDART00000177784
ENSDART00000122009 |
wdr59
|
WD repeat domain 59 |
| chr15_-_16704417 | 0.07 |
ENSDART00000155163
|
caln1
|
calneuron 1 |
| chr1_+_29267841 | 0.07 |
ENSDART00000085648
|
lig4
|
ligase IV, DNA, ATP-dependent |
| chr2_+_9970942 | 0.07 |
ENSDART00000153063
|
sec22ba
|
SEC22 homolog B, vesicle trafficking protein a |
| chr17_-_6534875 | 0.07 |
ENSDART00000156903
ENSDART00000155870 |
cenpo
|
centromere protein O |
| chr9_-_48296469 | 0.07 |
ENSDART00000058255
|
bbs5
|
Bardet-Biedl syndrome 5 |
| chr17_-_11439815 | 0.07 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr17_+_24623561 | 0.07 |
ENSDART00000156734
|
thumpd2
|
THUMP domain containing 2 |
| chr17_-_4252221 | 0.07 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr7_-_17337233 | 0.07 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr4_-_2637689 | 0.07 |
ENSDART00000192550
ENSDART00000021953 ENSDART00000150344 |
cog5
|
component of oligomeric golgi complex 5 |
| chr15_+_24572926 | 0.07 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr24_+_38301080 | 0.07 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr9_+_27379014 | 0.07 |
ENSDART00000167366
|
tlr20.4
|
toll-like receptor 20, tandem duplicate 4 |
| chr4_+_4889257 | 0.06 |
ENSDART00000175651
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_+_37760493 | 0.06 |
ENSDART00000012826
|
pex3
|
peroxisomal biogenesis factor 3 |
| chr6_-_24358732 | 0.06 |
ENSDART00000159595
|
ephx4
|
epoxide hydrolase 4 |
| chr18_-_14836600 | 0.06 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
Network of associatons between targets according to the STRING database.
First level regulatory network of hoxa2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 0.2 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.3 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 1.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.6 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.2 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.1 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.3 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.1 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.1 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.1 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.1 | GO:0032602 | chemokine production(GO:0032602) negative T cell selection(GO:0043383) thymocyte migration(GO:0072679) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.3 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.2 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.0 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.1 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.3 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.2 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.2 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.1 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.0 | 0.0 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |